Extending ebnm with custom ebnm-style functions
Source:vignettes/extending_ebnm.Rmd
extending_ebnm.RmdThe ebnm package, in addition to providing implementations of several commonly used priors (normal, Laplace, etc.), was designed to be easily extensible so that researchers are not limited by the existing options (despite the fact that a wide variety of options are available!).
In this vignette, we illustrate how to extend ebnm
by creating a custom EBNM solver in the style of other
ebnm functions such as ebnm_normal() and
ebnm_point_laplace(). Specifically, we implement an EBNM
solver, ebnm_t(), that uses the family of scaled
(Student’s) t priors. (As of this writing, this is not one of
the prior families included in ebnm.)
Please note: This vignette assumes that you have read the ebnm paper and are familiar with the basic functionality of the ebnm package.
The scaled-t prior family
The empirical Bayes normal means (EBNM) model with scaled-t prior is:
is the family of scaled-t priors, defined as follows:
where denotes the density function of the t distribution at with degrees of freedom. Fitting the prior therefore involves estimating two parameters: the scale parameter and the degrees of freedom .
Overview of the implementation process
The ebnm package is intended to encompass a very broad range of prior families. In general, creating a custom EBNM solver involves the following steps:
Define the prior family class .
Implement a function that estimates the prior .
Implement a function that computes summaries of the posteriors .
Create the main EBNM solver function.
Test the new EBNM solver.
Use the solver to analyze a data set.
In the following sections, we work through each of these steps in
detail with the aim of creating a new function ebnm_t()
that can fit the EBNM model with scaled-t prior.
For readability, we advise adhering to the Tidyverse style guide. Functions
should also be carefully tested; at minimum, functions should pass the
tests in ebnm_check_fn(). Additional unit tests are
strongly encouraged. The ebnm package implements a
large suite of unit tests using the testthat
package.
Step 1: Define the prior family class
First, we define a data structure for the priors in our prior family.
ebnm uses these structures in two ways: (1) to store
information about the fitted prior
(via the fitted_g field in the returned "ebnm"
object); (2) to initialize solutions (via the g_init
argument).
Sometimes, an existing data structure can be used. For example,
ebnm_normal(), ebnm_point_normal(),
ebnm_normal_scale_mixture(), and
ebnm_point_mass() all share the "normalmix"
class. For the scaled-t prior, we define a new class,
"tdist", that includes the scale and degrees of
freedom:
tdist <- function (scale, df) {
structure(data.frame(scale, df), class = "tdist")
}Step 2: Implement the optimization function
Next we implement a function for estimating the two parameters specifying the prior. Prior estimation is typically done by maximizing the likelihood. There are many approaches one might take to solve this optimization problem, and the best approach very much depends on context. For an excellent overview of the many R packages that can be used for numerical optimization, please see the CRAN task view on optimization.
Here, we use the L-BFGS-B method (implemented by the
optim() function). There are at least a couple of reasons
why we prefer using L-BFGS-B: (1) it doesn’t require installing any
additional packages; (2) it allows for bound constraints, which is
helpful since the two parameters in the prior both need to be positive.
Setting sensible upper and lower bounds can also help avoid numerical
issues. Here, we use the constraints
and
:
opt_t <- function (x, s, sigma_init, nu_init) {
optim(
par = c(sigma_init, nu_init),
fn = function (par) -llik_t(x, s, par[1], par[2]),
method = "L-BFGS-B",
lower = c(min(s)/10, 1),
upper = c(max(x), 1e3)
)
}Our optimization function opt_t() calls another
function, llik_t(), which isn’t yet implemented: this
function should give us the log likelihood at the current parameter
estimates. (Note that, since optim() seeks to minimize the
objective, we compute the negative log likelihood.)
Computing the log likelihood involves taking 1-d integrals, or 1-d convolutions, over the unknown means :
Since we do not have a convenient closed-form expression for these
integrals, we compute them numerically using the
integrate() function:
llik_t <- function (x, s, sigma, nu) {
lik_one_obs <- function (x, s) {
integrate(lik_times_prior, -Inf, Inf, x = x, s = s,
sigma = sigma, nu = nu)$value
}
vlik <- Vectorize(lik_one_obs)
return(sum(log(vlik(x, s))))
}
lik_times_prior <- function (theta, x, s, sigma, nu) {
dnorm(x - theta, sd = s) * dt(theta / sigma, df = nu) / sigma
}(Optional) Include gradients in the optimization
As we found empirically in our numerical experiments,
providing the gradient calculations to optim() can in some
cases greatly speed up the optimization. When implementing your own
custom EBNM solvers, you should consider providing gradients,
particularly when analytic expressions are available (either via pen and
paper or via automatic differentiation).
Gradients for the scaled-t priors turn out to be difficult
to obtain, but to illustrate how one might provide them, we estimate
gradients numerically using the grad() function from the
numDeriv package. We include this code for illustrative
purposes; since optim() also computes gradients
numerically, we do not expect this solution to provide any speedup.
opt_t <- function (x, s, sigma_init, nu_init) {
optim(
par = c(sigma_init, nu_init),
fn = function (par) -llik_t(x, s, par[1], par[2]),
gr = function (par) -grad_t(x, s, par[1], par[2]),
method = "L-BFGS-B",
lower = c(min(s)/10, 1),
upper = c(max(x), 1e3)
)
}The grad_t() function used above will estimate the
gradients numerically using numDeriv:
library(numDeriv)
grad_t <- function (x, s, sigma, nu) {
grad(function(par) llik_t(x, s, par[1], par[2]), c(sigma, nu))
}Using this version of the opt_t function should produce
very similar results to the implementation that does not include the
gradient.
Step 3: Implement the posterior summary function
Once we’ve estimated a prior , we can compute summary statistics (means, variances, etc.) from the posterior distributions.
From Bayes’ rule, the posterior distribution for the i-th unknown mean is
For this example, we compute three posterior statistics: the posterior mean, the posterior second moment, and the posterior standard deviation. This is all accomplished by a single function that returns a data frame containing the posterior statistics:
post_summary_t <- function (x, s, sigma, nu) {
samp <- post_sampler_t(x, s, sigma, nu, nsamp = 1000)
return(data.frame(
mean = colMeans(samp),
sd = apply(samp, 2, sd),
second_moment = apply(samp, 2, function (x) mean(x^2))
))
}The missing piece is a function post_sampler_t() that
draws random samples from the posteriors. While drawing independent
samples is difficult, we can easily design an MCMC scheme to
approximately draw samples from the posteriors. This is implemented
using the mcmc package (which you should install if you
haven’t already):
# install.packages("mcmc")
library(mcmc)
post_sampler_t <- function (x, s, sigma, nu, nsamp) {
sample_one_theta <- function (x_i, s_i) {
lpostdens <- function (theta) {
dt(theta/sigma, df = nu, log = TRUE) -
log(sigma) +
dnorm(x_i - theta, sd = s_i, log = TRUE)
}
metrop(lpostdens, initial = x_i, nbatch = nsamp)$batch
}
vsampler <- Vectorize(sample_one_theta)
return(vsampler(x, s))
}This is most certainly not the most efficient nor numerically stable way to perform these computations. But we do it this way here to keep the example simple.
Step 4: Put it all together
Having implemented the key computations for our new EBNM solver, we
will now incorporate these computations into a single function,
ebnm_t(), which accepts the same inputs as the solvers in
the ebnm package.
For simplicity, we ignore the output parameter and just
return all the results (data, posterior summaries, fitted prior, log
likelihood and posterior sampler). See help(ebnm) for
details about the expected structure of the return value.
Here’s the new function:
ebnm_t <- function (x,
s = 1,
mode = 0,
scale = "estimate",
g_init = NULL,
fix_g = FALSE,
output = ebnm_output_default(),
optmethod = NULL,
control = NULL) {
# Some basic argument checks.
if (mode != 0) {
stop("The mode of the t-prior must be fixed at zero.")
}
if (scale != "estimate") {
stop("The scale of the t-prior must be estimated rather than fixed ",
"at a particular value.")
}
# If g_init is provided, extract the parameters. Otherwise, provide
# reasonable initial estimates.
if (!is.null(g_init)) {
sigma_init <- g_init$scale
nu_init <- g_init$df
} else {
sigma_init <- sqrt(mean(x^2))
nu_init <- 4
}
# If g is fixed, use g_init. Otherwise optimize g.
if (fix_g) {
sigma <- sigma_init
nu <- nu_init
llik <- llik_t(x, s, sigma, nu)
} else {
opt_res <- opt_t(x, s, sigma_init, nu_init)
sigma <- opt_res$par[1]
nu <- opt_res$par[2]
llik <- -opt_res$value
}
# Prepare the final output.
retval <- structure(list(
data = data.frame(x = x, s = s),
posterior = post_summary_t(x, s, sigma, nu),
fitted_g = tdist(scale = sigma, df = nu),
log_likelihood = llik,
post_sampler = function (nsamp) post_sampler_t(x, s, sigma, nu, nsamp)
), class = c("list", "ebnm"))
return(retval)
}Step 5: Verify the EBNM function
ebnm provides a function,
ebnm_check_fn(), that runs basic tests to verify that the
EBNM function works as expected. Let’s run the checks using a small,
simulated data set:
Step 6: Use the new EBNM function to analyze a data set
Finally, we analyze a simulated data set in which the unobserved means are simulated from a t distribution with a scale of 2 and 5 degrees of freedom:
Let’s compare the use of the scaled-t prior with a normal prior:
normal_res <- ebnm_normal(x, s = 1)
t_res <- ebnm_t(x, s = 1)(Note that the call to ebnm_t() is considerably slower
than the call to ebnm_normal() because the computations
with the scaled-t prior are more complex and we did not put any
effort into making the computations efficient.)
Let’s compare the two results:
plot(normal_res, t_res)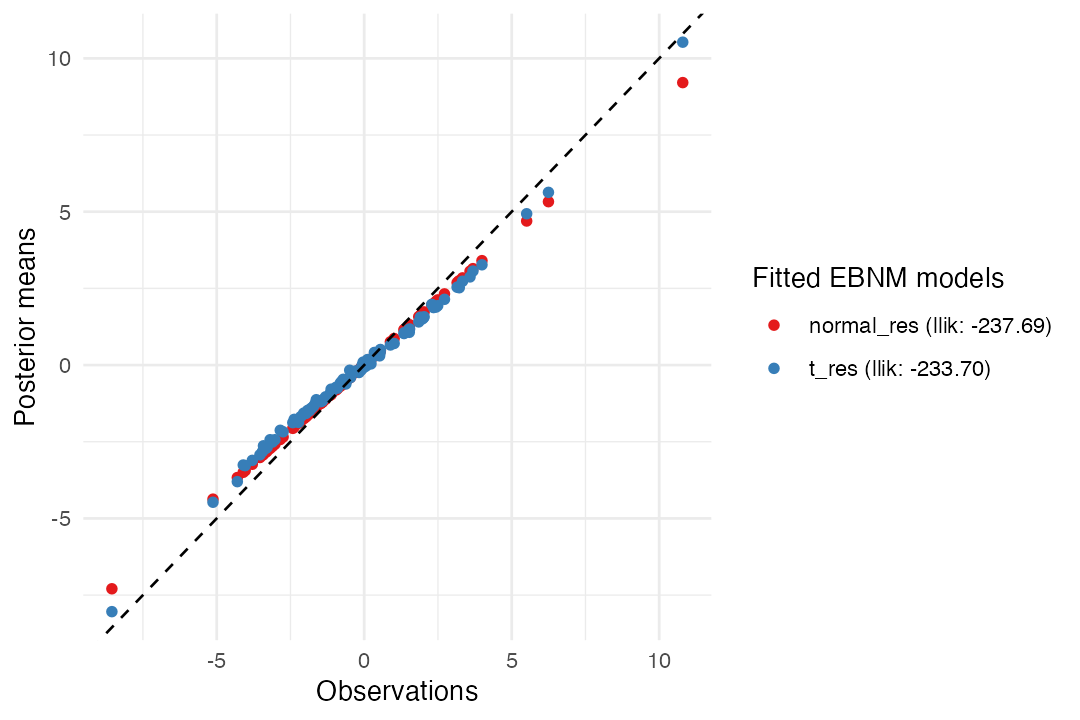
ebnm_t() shrinks large observations less aggressively
than ebnm_normal() and so the fit with the
scaled-t prior results in slightly more accurate estimates:
rmse_normal <- sqrt(mean((coef(normal_res) - theta)^2))
rmse_t <- sqrt(mean((coef(t_res) - theta)^2))
c(rmse_normal = rmse_normal, rmse_t = rmse_t)
# rmse_normal rmse_t
# 0.9056053 0.8662794Reassuringly, the parameters of the estimated prior are similar to the simulation parameters (, ):
c(t_res$fitted_g)
# $scale
# [1] 1.785927
#
# $df
# [1] 4.456856Session information
The following R version and packages were used to generate this vignette:
sessionInfo()
# R version 4.5.1 (2025-06-13)
# Platform: aarch64-apple-darwin20
# Running under: macOS Sequoia 15.5
#
# Matrix products: default
# BLAS: /Library/Frameworks/R.framework/Versions/4.5-arm64/Resources/lib/libRblas.0.dylib
# LAPACK: /Library/Frameworks/R.framework/Versions/4.5-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.1
#
# locale:
# [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#
# time zone: America/New_York
# tzcode source: internal
#
# attached base packages:
# [1] stats graphics grDevices utils datasets methods base
#
# other attached packages:
# [1] ebnm_1.1-38 mcmc_0.9-8
#
# loaded via a namespace (and not attached):
# [1] sass_0.4.10 generics_0.1.4 ashr_2.2-63 lattice_0.22-7
# [5] digest_0.6.37 magrittr_2.0.3 evaluate_1.0.4 grid_4.5.1
# [9] RColorBrewer_1.1-3 fastmap_1.2.0 jsonlite_2.0.0 Matrix_1.7-3
# [13] mixsqp_0.3-54 scales_1.4.0 truncnorm_1.0-9 invgamma_1.2
# [17] textshaping_1.0.1 jquerylib_0.1.4 cli_3.6.5 rlang_1.1.6
# [21] deconvolveR_1.2-1 splines_4.5.1 withr_3.0.2 cachem_1.1.0
# [25] yaml_2.3.10 tools_4.5.1 SQUAREM_2021.1 dplyr_1.1.4
# [29] ggplot2_3.5.2 vctrs_0.6.5 R6_2.6.1 lifecycle_1.0.4
# [33] fs_1.6.6 htmlwidgets_1.6.4 trust_0.1-8 ragg_1.4.0
# [37] irlba_2.3.5.1 pkgconfig_2.0.3 desc_1.4.3 pkgdown_2.1.3
# [41] bslib_0.9.0 pillar_1.11.0 gtable_0.3.6 glue_1.8.0
# [45] Rcpp_1.1.0 systemfonts_1.2.3 xfun_0.52 tibble_3.3.0
# [49] tidyselect_1.2.1 rstudioapi_0.17.1 knitr_1.50 farver_2.1.2
# [53] htmltools_0.5.8.1 labeling_0.4.3 rmarkdown_2.29 compiler_4.5.1
# [57] horseshoe_0.2.0