In this vignette, we demonstrate a procedure that helps SuSiE get out of local optimum.
We simulate phenotype using UK Biobank genotypes from 50,000 individuals. There are 1001 SNPs. It is simulated to have exactly 2 non-zero effects at 234, 287.
library(susieR)
library(curl)
# Using libcurl 8.18.0 with OpenSSL/3.6.1
data_file <- tempfile(fileext = ".RData")
data_url <- paste0("https://raw.githubusercontent.com/stephenslab/susieR/",
"master/inst/datafiles/FinemappingConvergence1k.RData")
curl_download(data_url,data_file)
load(data_file)
b <- FinemappingConvergence$true_coef
susie_plot(FinemappingConvergence$z, y = "z", b=b)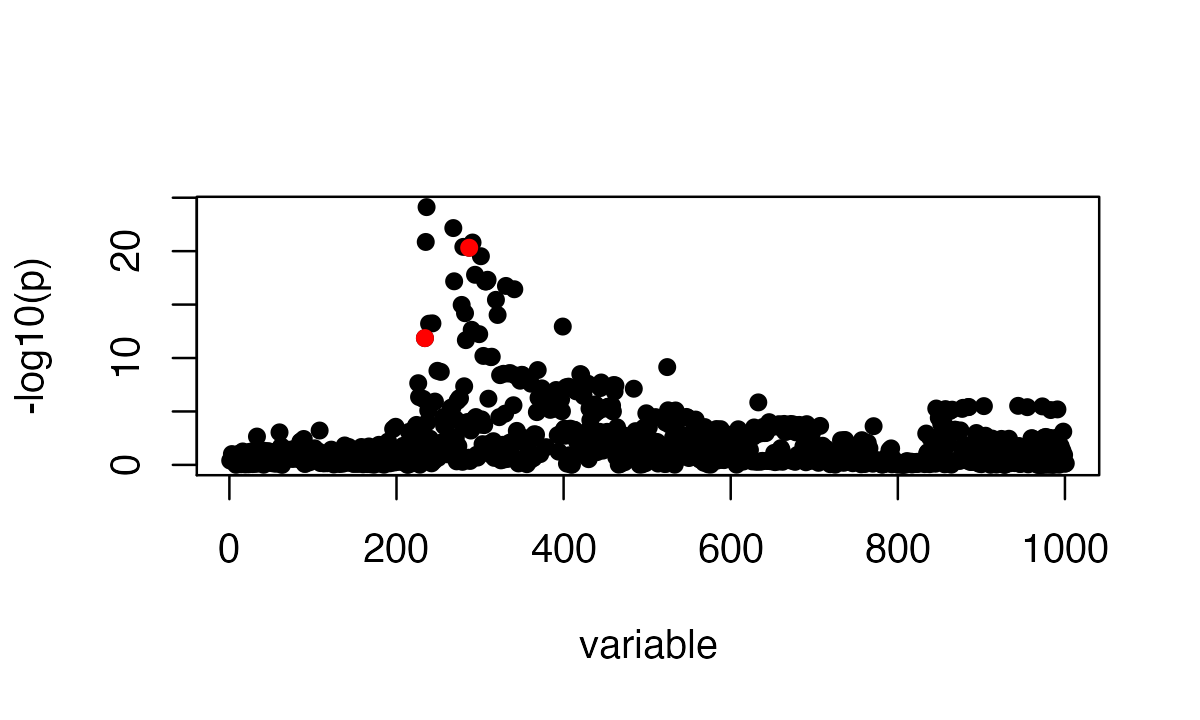
The strongest marginal association is a non-effect SNP.
Since the sample size is large, we use sufficient statistics (X^\intercal X, X^\intercal y, y^\intercal y
and sample size n) to fit susie model.
It identifies 2 Credible Sets, one of them is false positive. This is
because susieR get stuck around a local minimum.
fitted <- with(FinemappingConvergence,
susie_ss(XtX = XtX, Xty = Xty, yty = yty, n = n))
susie_plot(fitted, y="PIP", b=b, main=paste0("ELBO = ", round(susie_get_objective(fitted),2)))
Our refine procedure to get out of local optimum is
fit a susie model, s (suppose it has K CSs).
for CS in s, set SNPs in CS to have prior weight 0, fit susie model –> we have K susie models: t_1, \cdots, t_K.
for each k = 1, \cdots, K, fit susie with initialization at t_k (\alpha, \mu, \mu^2) –> s_k
if \max_k \text{elbo}(s_k) > \text{elbo}(s), set s = s_{kmax} where kmax = \arg_k \max \text{elbo}(s_k) and go to step 2; if no, break.
We fit susie model with above procedure by setting
refine = TRUE.
fitted_refine <- with(FinemappingConvergence,
susie_ss(XtX = XtX, Xty = Xty, yty = yty,
n = n, refine=TRUE))
susie_plot(fitted_refine, y="PIP", b=b, main=paste0("ELBO = ", round(susie_get_objective(fitted_refine),2)))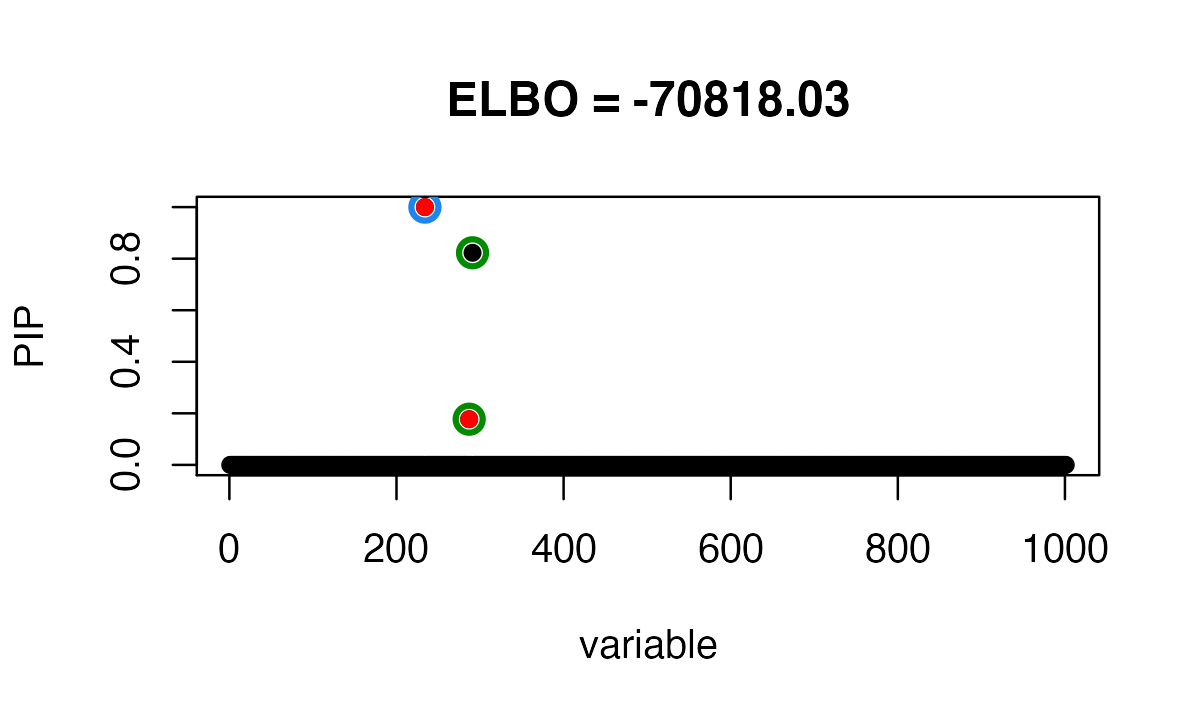
With the refine procedure, it identifies 2 CSs with the true signals, and the achieved evidence lower bound (ELBO) is higher.
Session information
Here are some details about the computing environment, including the versions of R, and the R packages, used to generate these results.
sessionInfo()
# R version 4.4.3 (2025-02-28)
# Platform: x86_64-conda-linux-gnu
# Running under: Ubuntu 24.04.3 LTS
#
# Matrix products: default
# BLAS/LAPACK: /home/runner/work/susieR/susieR/.pixi/envs/r44/lib/libopenblasp-r0.3.30.so; LAPACK version 3.12.0
#
# locale:
# [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
# [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
# [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
# [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#
# time zone: Etc/UTC
# tzcode source: system (glibc)
#
# attached base packages:
# [1] stats graphics grDevices utils datasets methods base
#
# other attached packages:
# [1] curl_7.0.0 susieR_0.15.51
#
# loaded via a namespace (and not attached):
# [1] Matrix_1.7-4 gtable_0.3.6 jsonlite_2.0.0
# [4] compiler_4.4.3 crayon_1.5.3 Rcpp_1.1.1
# [7] jquerylib_0.1.4 systemfonts_1.3.1 scales_1.4.0
# [10] textshaping_1.0.4 yaml_2.3.12 fastmap_1.2.0
# [13] lattice_0.22-9 plyr_1.8.9 ggplot2_4.0.2
# [16] R6_2.6.1 mixsqp_0.3-54 knitr_1.51
# [19] htmlwidgets_1.6.4 zigg_0.0.2 desc_1.4.3
# [22] bslib_0.10.0 RColorBrewer_1.1-3 rlang_1.1.7
# [25] cachem_1.1.0 reshape_0.8.10 xfun_0.56
# [28] fs_1.6.6 sass_0.4.10 S7_0.2.1
# [31] RcppParallel_5.1.11-1 otel_0.2.0 cli_3.6.5
# [34] pkgdown_2.2.0 digest_0.6.39 grid_4.4.3
# [37] irlba_2.3.7 lifecycle_1.0.5 vctrs_0.7.1
# [40] Rfast_2.1.5.2 evaluate_1.0.5 glue_1.8.0
# [43] farver_2.1.2 ragg_1.5.0 rmarkdown_2.30
# [46] matrixStats_1.5.0 tools_4.4.3 htmltools_0.5.9