Illustration of the limitations of SuSiE for molecular phenotype data
William R.P. Denault
2024-03-07
Source:vignettes/Limitation_SuSiE.Rmd
Limitation_SuSiE.RmdThis vignette illustrates some of the limitations of SuSiE when analyzing molecular traits or traits/responses variables that are functions. We use a simple example where the effect of a SNP spans over multiple CpG. We start by fine-mapping the effect of the genotype on each CpG in a sequential fashion. Then, we show that SuSiE can output inconsistent results. Finally we show that fSuSiE can handle this problem and output much more interpretable results.
Let’s simulate a simple effect of a SNP on a set of CpGs. The effect of the SNP is a cosine function that spans over 40 CpGs.
effect <- 1.2 * cos((1:128)/128 * 3*pi)
effect[which(effect < 0)] <- 0
effect[1:40] <- 0
plot(effect,pch = 19)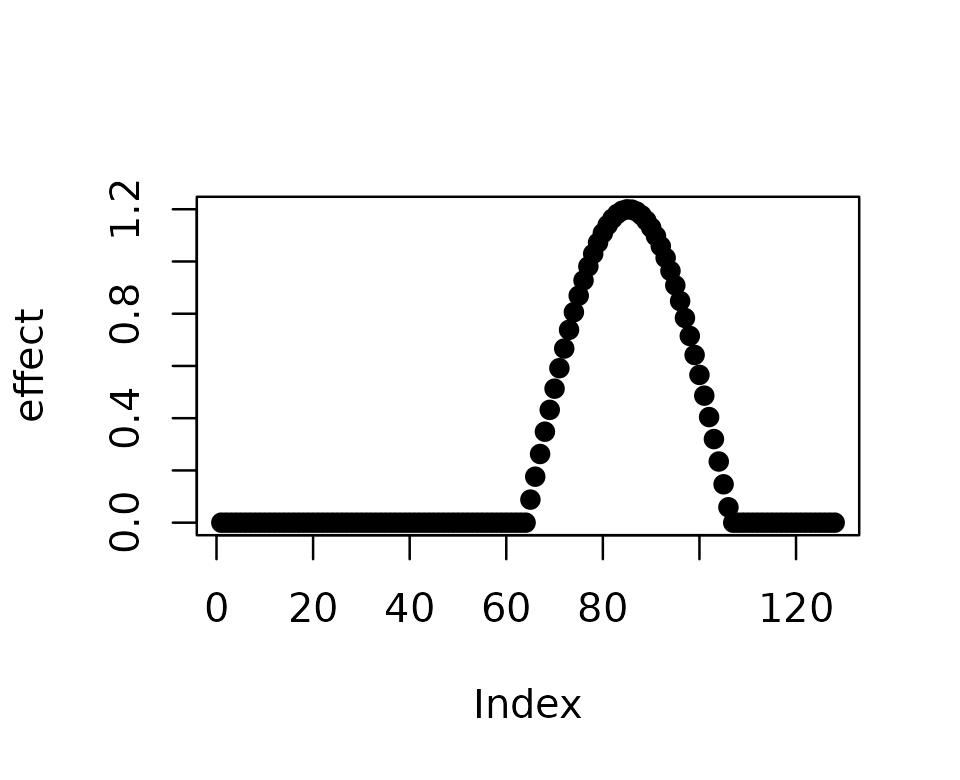
We simulate 100 observations of the effect of the SNP on the CpG.
set.seed(1)
data(N3finemapping)
X <- N3finemapping$X[1:100,]
y <- matrix(0,100,128)
true_pos <- 700
for (i in 1:100)
y[i,] <- X[i,true_pos] * effect + rnorm(length(effect))
which(effect > 0)
# [1] 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83
# [20] 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102
# [39] 103 104 105 106Once the data is simulated, we can run SuSiE on each CpG. For the sake of simplicity, we only run SuSiE with L = 1 as we are aware that there is only one effect in this simulation. Also for simplicity, we assume that the association analysis has been performed with 100% accuracy—that is, we know exactly which CpGs are the affected CpGs, and therefore we can ignore the others.
susie_est <- vector("list",128)
for (i in which(effect > 0))
susie_est[[i]] <- susie(X,y[,i],L = 1)$sets
names(susie_est) <- paste0("cpg",1:128)
susie_est <- susie_est[effect > 0]One issue is that SuSiE fails to detect a causal SNP at most of the affected CpGs:
sapply(susie_est,function (x) length(x$cs))
# cpg65 cpg66 cpg67 cpg68 cpg69 cpg70 cpg71 cpg72 cpg73 cpg74 cpg75
# 0 0 0 0 0 0 0 0 0 0 0
# cpg76 cpg77 cpg78 cpg79 cpg80 cpg81 cpg82 cpg83 cpg84 cpg85 cpg86
# 0 0 0 1 0 0 0 0 0 0 0
# cpg87 cpg88 cpg89 cpg90 cpg91 cpg92 cpg93 cpg94 cpg95 cpg96 cpg97
# 0 1 0 0 1 0 0 1 0 0 0
# cpg98 cpg99 cpg100 cpg101 cpg102 cpg103 cpg104 cpg105 cpg106
# 0 0 1 0 0 0 0 0 0A second issue is that the SuSiE produces multiple fine-mapping signals (and therefore multiple sets of CSs), so it is unclear how to reason about uncertainty in the causal SNP(s). For example, compare the SuSiE results for CpGs 88 and 91:
df_effect <- data.frame(x = 1:128,y = effect)
P00 <- ggplot(df_effect,aes(x = x,y = y)) +
geom_point() +
ylim(c(-0.1,1.3)) +
theme_classic() +
theme(panel.border = element_rect(color = "black",fill = NA,linewidth = 1.2),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank())
P01 <- P00 +
geom_point(x = 88,y = effect[88],color = "red",shape = 21,size = 3) +
geom_hline(yintercept = 0) +
labs(x = "CpG",y = "effect",title = "CpG 88")
tt <- susie(X,y[,88],L = 1)
df_pip1 <- data.frame(x = 1:length(tt$pip),y = tt$pip)
df2 <- data.frame(x = tt$sets$cs$L1,y = tt$pip[tt$sets$cs$L1])
df3 <- data.frame(x = 700,y = tt$pip[700])
P11 <- ggplot() +
geom_point(df_pip1,mapping = aes(x = x,y = y)) +
labs(x = "SNP",y = "PIP",title = "CpG 88") +
theme_classic() +
theme(panel.border = element_rect(color = "black",fill = NA,linewidth = 1.2),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank())+
geom_point(df2,mapping = aes(x = x,y = y),color = "dodgerblue",size = 3)+
geom_point(df3,mapping = aes(x = x,y = y),color = "red",shape = 21,size = 3)
P02 <- P00 +
geom_point(x = 91,y = effect[91],color = "red",shape = 21,size = 3) +
labs(x = "CpG",y = "effect",title = "CpG 91") +
geom_hline(yintercept = 0) +
theme(axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank())
tt <- susie(X,y[,91],L = 1)
df_pip2 <- data.frame(x = 1:length(tt$pip),y = tt$pip)
df2 <- data.frame(x = tt$sets$cs$L1,y = tt$pip[tt$sets$cs$L1])
df3 <- data.frame(x = 700,y = tt$pip[700])
P21 <- ggplot() +
geom_point(df_pip2,mapping = aes(x = x,y = y)) +
labs(x = "SNP",y = "PIP",title = "CpG 91") +
theme_classic() +
theme(panel.border = element_rect(color = "black",fill = NA,linewidth = 1.2),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank()) +
geom_point(df2,mapping = aes(x = x,y = y),color = "dodgerblue",size = 3) +
geom_point(df3,mapping = aes(x = x,y = y),color = "red",shape = 21,size = 3)
P03 <- P00 +
geom_point(x = 94,y = effect[94],color = "red",shape = 21,size = 3)+
theme_classic() +
theme(panel.border = element_rect(color = "black",fill = NA,linewidth = 1.2),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank()) +
geom_hline(yintercept = 0) +
labs(x = "CpG",y = "effect",title = "CpG 91")
plot_grid(P01,P11,
P02,P21,
nrow = 2,ncol = 2)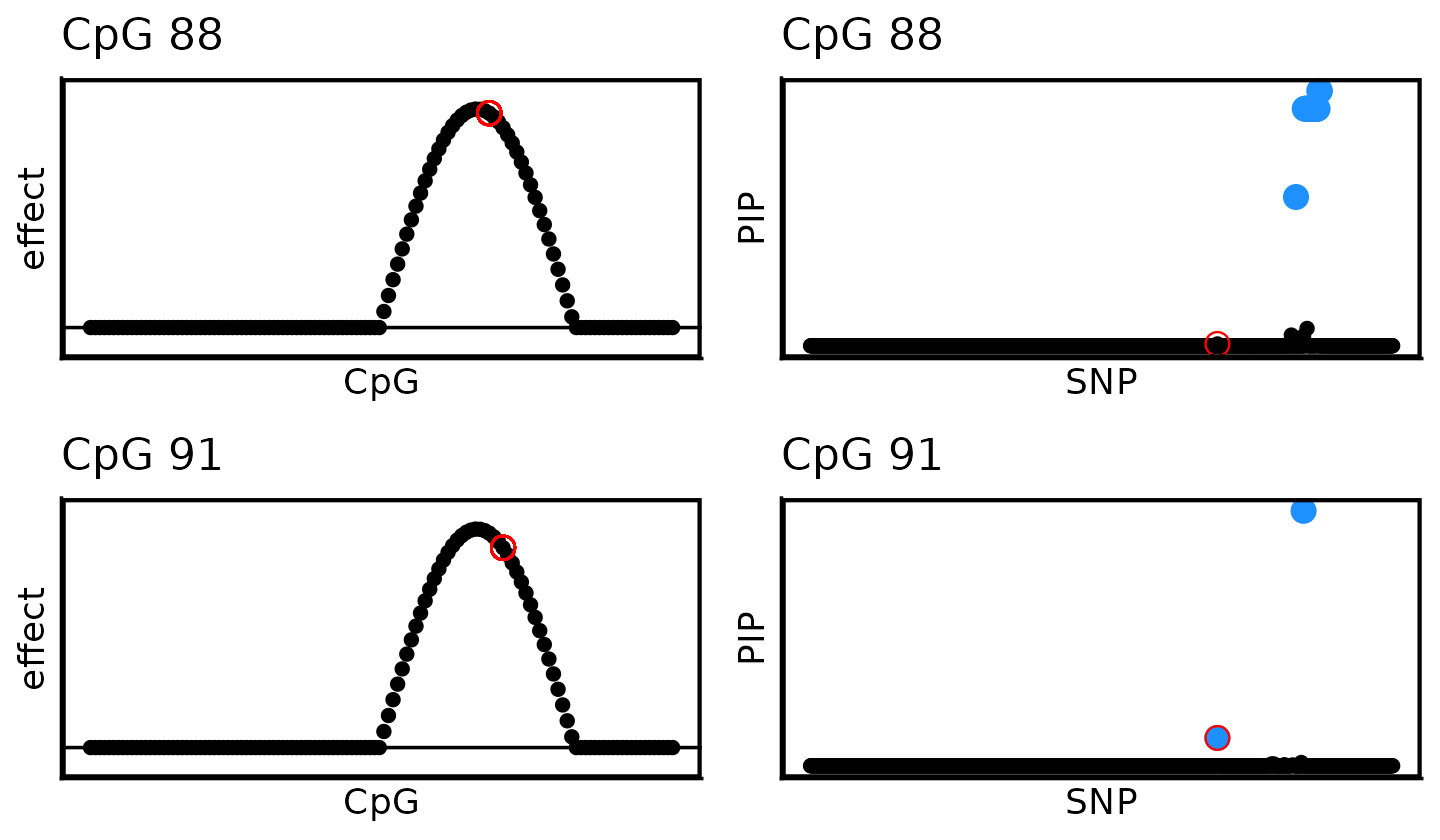
Now compare to an analysis of these data using fSuSiE, which involves a single call to the “susiF” function:
tt <- susiF(y,X,L = 1,post_processing = "smash",verbose = FALSE)
tt$cs
# [1] "Fine mapping done, refining effect estimates using cylce spinning wavelet transform"
# [[1]]
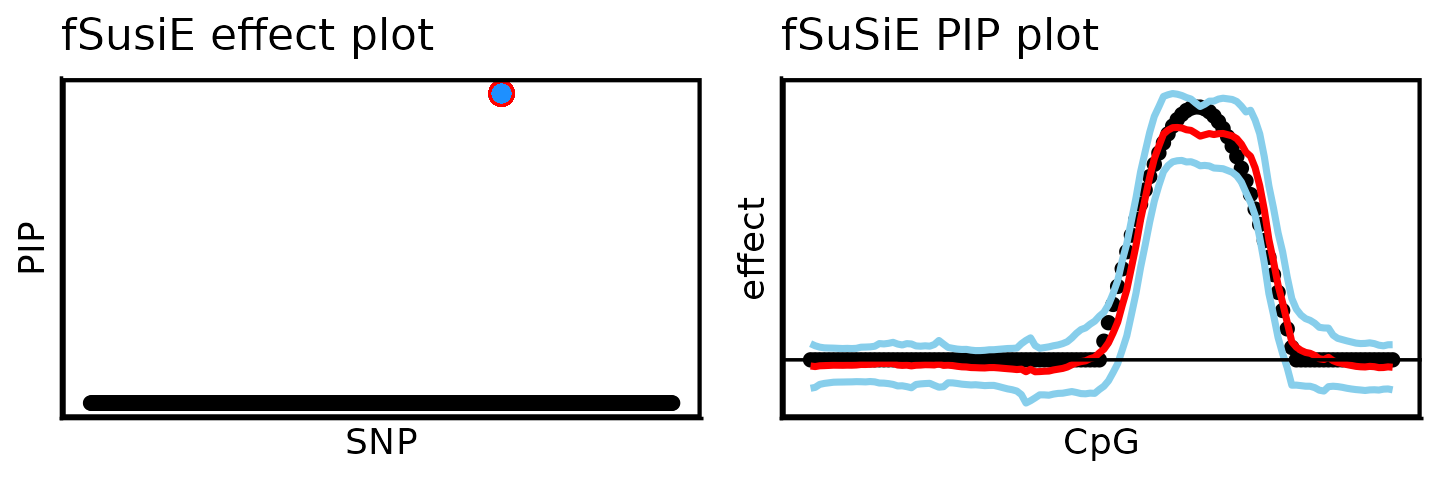
# [1] 691Notice that fSuSiE returned a single CS containing 1 SNP, which is the true causal SNP.
These plots show two other results: on the left-hand side, the fine-mapping signal (PIPs); on the right-hand side, the estimated effect on the methylation levels (red = posterior mean, light blue = credible bands).
tt$pip <- rep( 0,length(tt$pip))
tt$pip[700] <-1
df_pip5 <-data.frame(x=1:length(tt$pip),
y=tt$pip)
df_est_f<- data.frame(y = c(tt$fitted_func[[1]],
tt$cred_band[[1]][1,],
tt$cred_band[[1]][2,]),
x = rep(1:128,3),
type = factor(rep(1:3,each = 128)))
df_est_f <- data.frame(y = c(tt$fitted_func[[1]],
tt$cred_band[[1]][1,],
tt$cred_band[[1]][2,]),
x = rep(1:128,3),
type = factor(rep(1:3,each = 128)))
P50 <- ggplot() +
geom_point(df_effect,mapping = aes(x = x,y = y)) +
geom_line(df_est_f[which(df_est_f$type == 1),],
mapping = aes(x = x,y = y,linetype = "dashed"),
color = "red",linewidth = 1) +
geom_line(df_est_f[which(df_est_f$type == 2),],
mapping = aes(x = x,y = y),
color = "skyblue",linewidth = 1)+
geom_line(df_est_f[which(df_est_f$type == 3),],
mapping=aes(x = x,y = y),
color = "skyblue",linewidth = 1) +
labs(x = "CpG",y = "effect",title = "fSuSiE PIP plot") +
theme_classic() +
geom_hline(yintercept = 0) +
theme(legend.position = "none",
panel.border = element_rect(color = "black",fill = NA,linewidth = 1.2),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank())
P51 <- ggplot(df_pip5,aes(x = x,y = y))+
geom_point() +
labs(x = "SNP",y = "PIP",title = "fSusiE effect plot") +
theme_classic() +
geom_point(x = 700,y = tt$pip[700],color = "dodgerblue",size = 3)+
theme(panel.border = element_rect(color = "black",fill = NA,linewidth = 1.2),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank())+
geom_point(x = 700,y = tt$pip[700],color = "red",shape = 21,size = 3)
plot_grid(P51,P50,nrow = 1,ncol = 2)