An R example: ashr benchmark
This is a more advanced application of DSC with R scripts. We demonstrate in this tutorial features of DSC including:
- Inline code as parameters
@ALIASdecorator- R library installation and version check
- Use
dscrutilsto query results limited to given conditions
DSC Problem
The DSC problem is based on the ASH example of DSCR (R Markdown version and HTML version). Material to run this tutorial can be found in DSC vignettes repo. Description below is copied from the DSCR vignette:
To illustrate we consider the problem of shrinkage, which is tackled by the
ashrpackage at http://www.github.com/stephens999/ashr. The input to this DSC is a set of estimates $\hat\beta$, with associated standard errors $s$. These values are estimates of actual (true) values for $\beta$, so the meta-data in this case are the true values of beta. Methods must take $\hat\beta$ and $s$ as input, and provide as output “shrunk” estimates for $\beta$ (so output is a list with one element, calledbeta_est, which is a vector of estimates for beta). The score function then scores methods on their RMSE comparingbeta_estwith beta.
First define a datamaker which simulates true values of $\beta$ from a user-specified normal mixture, where one of the components is a point mass at 0 of mass $\pi_0$, which is a user-specified parameter. It then simulates $\hat\beta \sim N(\beta_j,s_j)$ (where $s_j$ is again user-specified). It returns the true $\beta$ values and true $\pi_0$ value as meta-data, and the estimates $\hat\beta$ and $s$ as input-data.
Now define a method wrapper for the
ashfunction from theashrpackage. Notice that this wrapper does not return output in the required format - it simply returns the entire ash output.
Finally add a generic (can be used to deal with both $\pi$ and $\beta$) score function to evaluate estimates by
ash.
DSC Specification
The problem is fully specified in DSC syntax below, following the structure of the original DSCR implementation:
#!/usr/bin/env dsc
simulate: datamaker.R
# module input and variables
mixcompdist: normalmix
g: (c(2/3,1/3),c(0,0),c(1,2)),
(rep(1/7,7),c(-1.5,-1,-0.5,0,0.5,1,1.5),rep(0.5,7)),
(c(1/4,1/4,1/3,1/6),c(-2,-1,0,1),c(2,1.5,1,1))
min_pi0: 0
max_pi0: 1
nsamp: 1000
betahatsd: 1
# module decoration
@ALIAS: args = List()
# module output
$data: data
$true_beta: data$meta$beta
$true_pi0: data$meta$pi0
shrink: runash.R
# module input and variables
data: $data
mixcompdist: "normal", "halfuniform"
# module output
$ash_data: ash_data
$beta_est: ashr::get_pm(ash_data)
$pi0_est: ashr::get_pi0(ash_data)
score_beta: score.R
# module input and variables
est: $true_beta
truth: $beta_est
# module output aka pipeline variable
$mse: result
score_pi0: score.R
# module input and variables
est: $pi0_est
truth: $true_pi0
# module output
$mse: result
DSC:
# module ensembles
define:
score: score_beta, score_pi0
# pipelines
run: simulate * shrink * score
# runtime environments
R_libs: ashr@stephens999 (2.2.7+)
exec_path: bin
output: dsc_result
It is suggested that you check out the corresponding R codes for modules simulate, shrink and the score function to figure out how DSC communicates with your R scripts.
Here we will walk through all modules to highlight important syntactical elements.
Module simulate
Un-quoted string as parameters and output values
The parameter g has three candidate values, all of which are un-quoted string to be plugged to script as is. In other words, DSC will interpret each as, eg, g <- list(c(2/3,1/3),c(0,0),c(1,2)) so that g will be assigned at runtime the output of R codes as specified for use with datamaker.R. Similarly, mixcompdist: normalmix will be interpreted as mixcompdist <- normalmix – this statement is valid as long as datamaker.R has loaded ashr library, because normalmix is function from ashr library.
Decorator @ALIAS for R list
Inside datamaker.R the input for the core function is a single parameter of an R list containing all parameters specified in this module. The decorator @ALIAS uses a special DSC operation List() to consolidate these parameters into an R list args which corresponds to the input parameter in datamaker.R.
Module shrink
Here notice the output variable are also provided at runtime: DSC allows output variables to directly accept pieces of codes that can provide the desired output. In this case, get_pi0 function from ashr package will get us what we need.
Module beta_score & pi0_score
These modules uses the same module executable score.R but on different input data. Due to differences in variable names it makes sense to configure them in separate blocks. However an alternative style that configures them in the same block is:
score_beta, score_pi0: score.R
@score_beta:
est: $true_beta
truth: $beta_est
$mse_beta: result
@score_pi0:
est: $pi0_est
truth: $true_pi0
$mse_pi: result
Here @* are module specific variable decorations that configures input and output such that different modules can be allowed in the same block.
Notice that different from the DSCR ASH example the output score is an “atomic” value (a float number). If the outcome object result is not such a simple object, for example it returns an R list, then you may want to code it to only extract the information you need so that they’ll be readily extractable in the benchmark query process – the query process can only extract atomic types. To do so, you can write something like, e.g., $mse_pi: score_output$mse.
DSC section for benchmark configuration
As has been discussed in previous tutorials, DSC sections defines module / pipeline ensembles and a benchmark. This DSC executes essentially two pipelines (one ending with score_beta another with score_pi0). The R_libs property specifies the R package required by the DSC. It is formatted as a github package (pkg@repo) and the minimal version requirement is 2.0.0. DSC will check first if the package is available and up-to-date, and install it if necessary. It will then check its version and quit on error if it does not satisfy the requirement. For safety concerns, DSC does not attempt to upgrade/downgrade a package in cases of version mismatch.
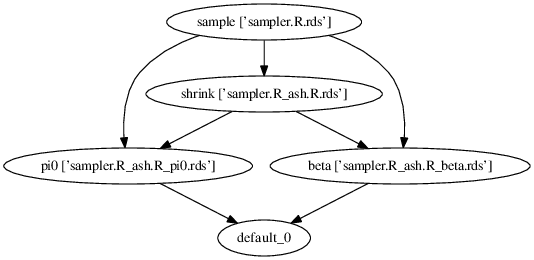
Execution logic
This diagram (generated by dot command using execution graph of this DSC) shows the logic of the benchmark:
Run DSC
We now run this DSC with 10 replicates:
cd ~/GIT/dsc/vignettes/ash
./settings.dsc --replicate 10
INFO: Checking R library ashr@stephens999/ashr ...
INFO: Checking R library dscrutils@stephenslab/dsc/dscrutils ...
INFO: DSC script exported to dsc_result.html
INFO: Constructing DSC from ./settings.dsc ...
INFO: Building execution graph & running DSC ...
[#########] 9 steps processed (215 jobs completed)
INFO: Building DSC database ...
INFO: DSC complete!
INFO: Elapsed time 85.268 seconds.
Benchmark evaluation
Extract result of interest
The DSCR ASH example has names to various simulation settings and methods. Here we use dscrutils to reproduce the DSCR example. It is similar to what we have done in the DSC Introduction example, but we will demonstrate the use of condition argument, to create 3 data-sets:
setwd('~/GIT/dsc/vignettes/ash')
dsc_dir = 'dsc_result'
An = dscrutils::dscquery(dsc_dir, targets = c("simulate.true_pi0",
"shrink.mixcompdist",
"shrink.beta_est",
"shrink.pi0_est",
"score.mse"),
condition = "simulate.g = 'list(c(2/3,1/3),c(0,0),c(1,2))'")
Bn = dscrutils::dscquery(dsc_dir, targets = c("simulate.true_pi0",
"shrink.mixcompdist",
"shrink.beta_est",
"shrink.pi0_est",
"score.mse"),
condition = "simulate.g = 'list(rep(1/7,7),c(-1.5,-1,-0.5,0,0.5,1,1.5),rep(0.5,7))'")
Cn = dscrutils::dscquery(dsc_dir, targets = c("simulate.true_pi0",
"shrink.mixcompdist",
"shrink.beta_est",
"shrink.pi0_est",
"score.mse"),
condition = "simulate.g = 'list(c(1/4,1/4,1/3,1/6),c(-2,-1,0,1),c(2,1.5,1,1))'")
To view one of the resulting data frame:
head(An)
Explore results of interest
Suppose we are interested in comparing $\pi_0$ estimates under scenario An, for two flavors of shrink module normal and halfuniform.
normal = subset(An, shrink.mixcompdist == 'normal')
hu = subset(An, shrink.mixcompdist == 'halfuniform')
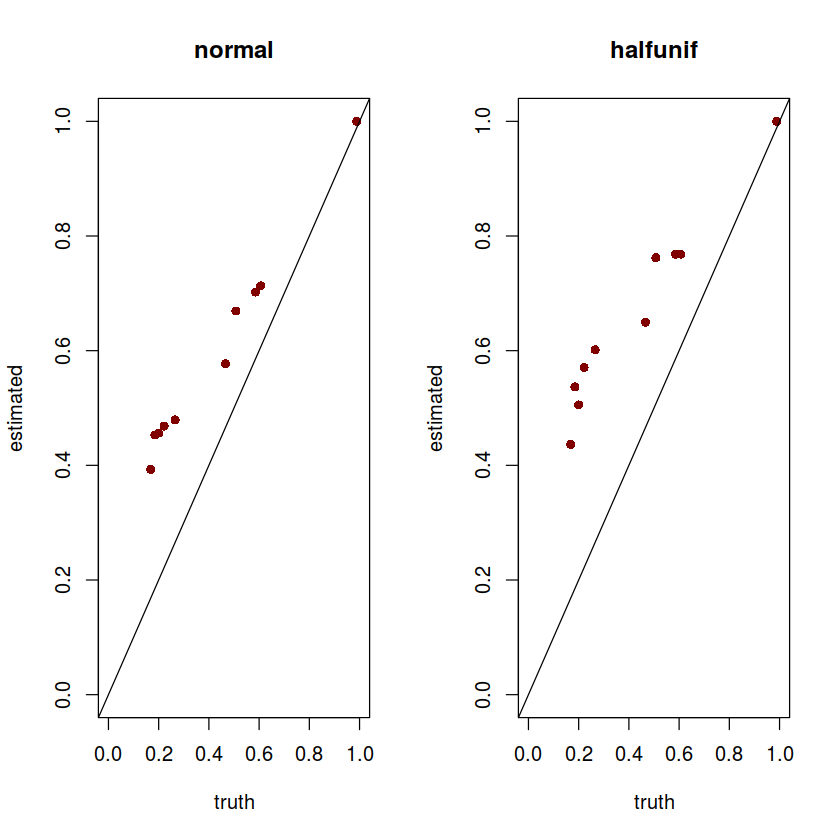
First, we visualize estimated vs true $\pi_0$ for two methods:
par(mfrow=c(1,2))
plot(subset(normal, score == 'score_pi0')$simulate.true_pi0,
subset(normal, score == 'score_pi0')$shrink.pi0_est,
xlab = 'truth', ylab = 'estimated', main = 'normal',
pch = 16, col = "#800000", xlim = c(0,1), ylim = c(0,1))
abline(a=0,b=1)
plot(subset(hu, score == 'score_pi0')$simulate.true_pi0,
subset(hu, score == 'score_pi0')$shrink.pi0_est,
xlab = 'truth', ylab = 'estimated', main = 'halfunif',
pch = 16, col = "#800000", xlim = c(0,1), ylim = c(0,1))
abline(a=0,b=1)
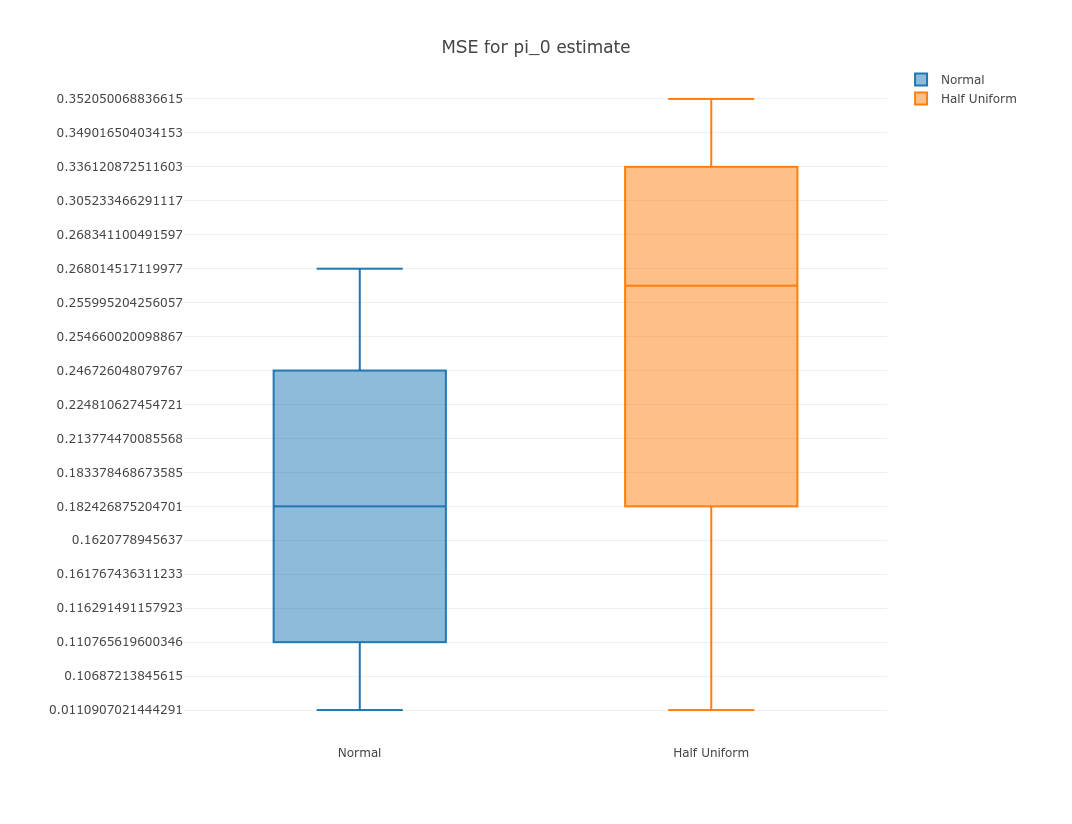
We then summarize the MSE scores for the two methods:
suppressMessages(library(plotly))
options(warn=-1)
p = plot_ly(y = subset(normal, score == 'score_pi0')$score.mse, name = 'Normal', type = "box") %>%
add_trace(y = subset(hu, score == 'score_pi0')$score.mse, name = 'Half Uniform', type = "box") %>%
layout(title = "MSE for pi_0 estimate")
export(p, file = 'mse.png')
IRdisplay::display_png(file="mse.png")
Explore intermediate output
We have previously focused on benchmark “results”, ie, the mean square errors. It is also possible to extract quantities of interest from any module in the benchmark. For example we want to explore relationship between “true” value and posterior estimates of $\beta$ from method normal and halfnormal. First we use dscrutils to extract these quantities:
dat2 = dscrutils::dscquery(dsc_dir,
targets = c("simulate.true_beta", "shrink.mixcompdist", "shrink.beta_est"),
condition = "simulate.g = 'list(c(2/3,1/3),c(0,0),c(1,2))'",
max.extract.vector = 1000)
print(dim(dat2))
head(dat2[,1:5])
As shown above, with max.extract.vector = 1000 we allow for extracting vector-valued $\beta$ estimates into the outputted data frame. The resulting dataframe has 2001 columns containing both $\beta$ and its posterior estimate $\tilde{\beta}{\text{normal}}$ and $\tilde{\beta}{\text{halfunif}}$. We make them into separate tables. Rows in each table are replicates.
pattern = "simulate.true_beta"
cols = which(sapply(strsplit(names(dat2),"[.]"), function (x) paste0(x[1],'.', x[2])) == pattern)
normal0 = subset(dat2, shrink.mixcompdist == 'normal')[,cols]
hu0 = subset(dat2, shrink.mixcompdist == 'halfuniform')[,cols]
#
pattern = "shrink.beta_est"
cols = which(sapply(strsplit(names(dat2),"[.]"), function (x) paste0(x[1],'.', x[2])) == pattern)
normal = subset(dat2, shrink.mixcompdist == 'normal')[,cols]
hu = subset(dat2, shrink.mixcompdist == 'halfuniform')[,cols]
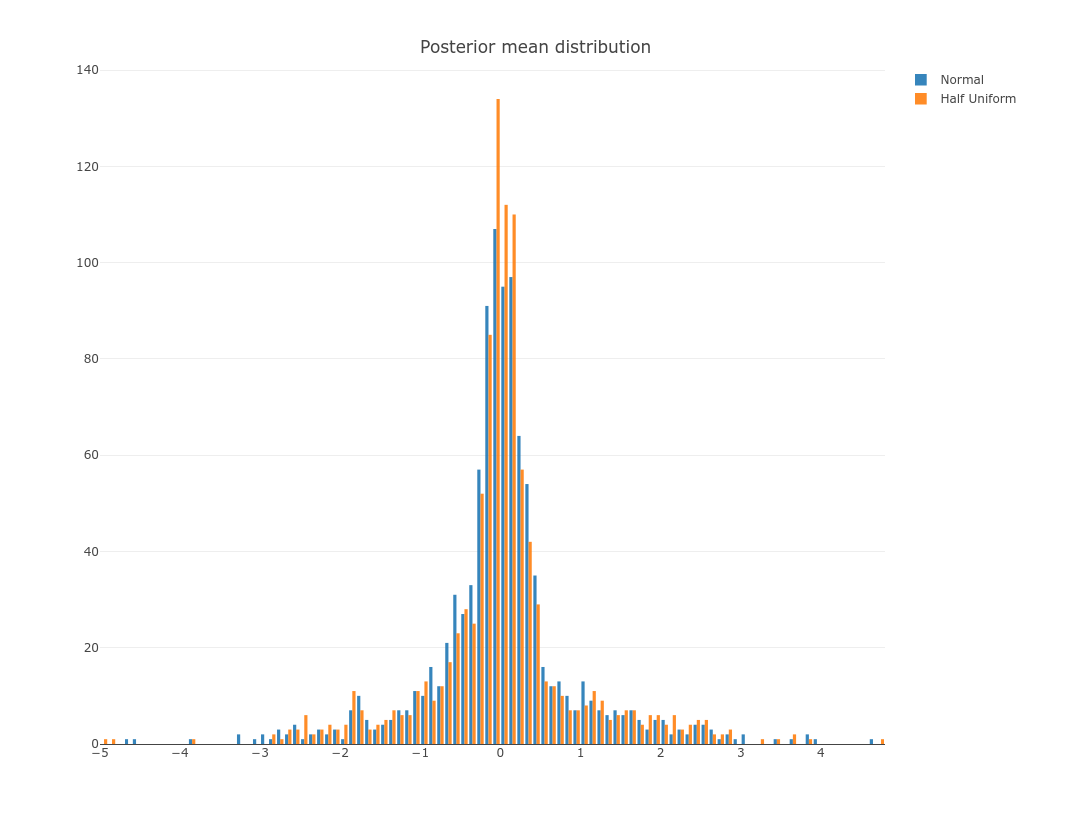
To view distribution of $\tilde{\beta}$ from one replicate:
p = plot_ly(x = as.numeric(normal[1,]), name = 'Normal', opacity = 0.9, type = "histogram") %>%
add_trace(x = as.numeric(hu[1,]), name = 'Half Uniform', opacity = 0.9, type = "histogram") %>%
layout(title = "Posterior mean distribution")
export(p, file = 'beta.png')
IRdisplay::display_png(file="beta.png")
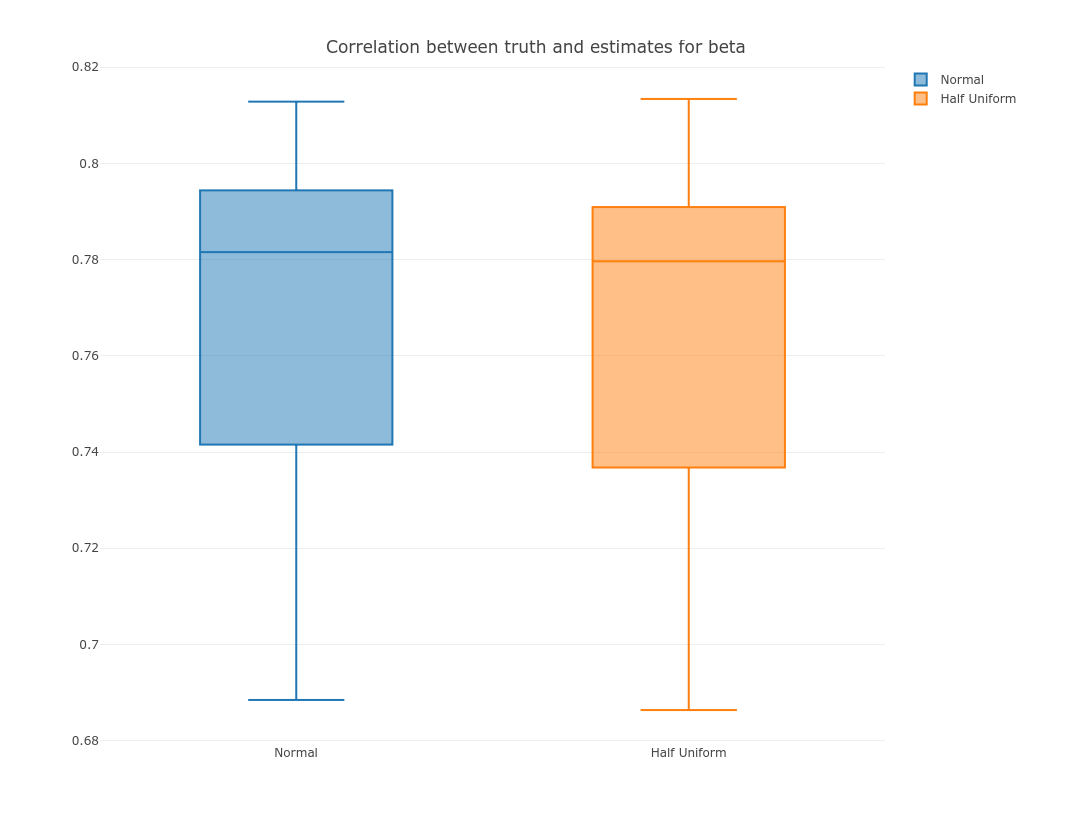
Now we compare correlation between estimate and the truth for these 2 methods:
normal0 = as.matrix(normal0)
normal = as.matrix(normal)
normal_cor = sapply(seq.int(dim(normal0)[1]), function(i) cor(normal0[i,], normal[i,]))
hu0 = as.matrix(hu0)
hu = as.matrix(hu)
hu_cor = sapply(seq.int(dim(hu0)[1]), function(i) cor(hu0[i,], hu[i,]))
p = plot_ly(y = normal_cor, name = 'Normal', type = "box") %>%
add_trace(y = hu_cor, name = 'Half Uniform', type = "box") %>%
layout(title = "Correlation between truth and estimates for beta")
export(p, file = 'corr.png')
IRdisplay::display_png(file="corr.png")