Fit Non-negative Matrix Factorization to Count Data
Source:R/fit_poisson_nmf.R, R/init_poisson_nmf.R
fit_poisson_nmf.RdApproximate the input matrix X by the
non-negative matrix factorization tcrossprod(L,F), in which
the quality of the approximation is measured by a
“divergence” criterion; equivalently, optimize the
likelihood under a Poisson model of the count data, X, in
which the Poisson rates are given by tcrossprod(L,F).
Function fit_poisson_nmf runs a specified number of
coordinate-wise updates to fit the L and F matrices.
fit_poisson_nmf(
X,
k,
fit0,
numiter = 100,
update.factors = seq(1, ncol(X)),
update.loadings = seq(1, nrow(X)),
method = c("scd", "em", "mu", "ccd"),
init.method = c("topicscore", "random"),
control = list(),
verbose = c("progressbar", "detailed", "none")
)
fit_poisson_nmf_control_default()
init_poisson_nmf(
X,
F,
L,
k,
init.method = c("topicscore", "random"),
beta = 0.5,
betamax = 0.99,
control = list(),
verbose = c("detailed", "none")
)
init_poisson_nmf_from_clustering(X, clusters, ...)Arguments
- X
The n x m matrix of counts; all entries of X should be non-negative. It can be a sparse matrix (class
"dgCMatrix") or dense matrix (class"matrix"), with some exceptions (see ‘Details’).- k
An integer 2 or greater giving the matrix rank. This argument should only be specified if the initial fit (
fit0orF, L) is not provided.- fit0
The initial model fit. It should be an object of class “poisson_nmf_fit”, such as an output from
init_poisson_nmf, or from a previous call tofit_poisson_nmf.- numiter
The maximum number of updates of the factors and loadings to perform.
- update.factors
A numeric vector specifying which factors (rows of
F) to update. By default, all factors are updated. Note that the rows that are not updated may still change by rescaling. WhenNULL, all factors are fixed. This option is only implemented formethod = "em"andmethod = "scd". If another method is selected, the default setting ofupdate.factorsmust be used.- update.loadings
A numeric vector specifying which loadings (rows of
L) to update. By default, all loadings are updated. Note that the rows that are not updated may still change by rescaling. WhenNULL, all loadings are fixed. This option is only implemented formethod = "em"andmethod = "scd". If another method is selected, the default setting ofupdate.loadingsmust be used.- method
The method to use for updating the factors and loadings. Four methods are implemented: multiplicative updates,
method = "mu"; expectation maximization (EM),method = "em"; sequential co-ordinate descent (SCD),method = "scd"; and cyclic co-ordinate descent (CCD),method = "ccd". See ‘Details’ for a detailed description of these methods.- init.method
The method used to initialize the factors and loadings. When
init.method = "random", the factors and loadings are initialized uniformly at random; wheninit.method = "topicscore", the factors are initialized using the (very fast) Topic SCORE algorithm (Ke & Wang, 2017), and the loadings are initialized by running a small number of SCD updates. This input argument is ignored if initial estimates of the factors and loadings are already provided via inputfit0, or inputsFandL.- control
A list of parameters controlling the behaviour of the optimization algorithm (and the Topic SCORE algorithm if it is used to initialize the model parameters). See ‘Details’.
- verbose
When
verbose = "detailed", information about the algorithm's progress is printed to the console at each iteration; whenverbose = "progressbar", a progress bar is shown; and whenverbose = "none", no progress information is printed. See the description of the “progress” return value for an explanation ofverbose = "detailed"console output. (Note that some columns of the “progress” data frame are not shown in the console output.)- F
An optional argument giving is the initial estimate of the factors (also known as “basis vectors”). It should be an m x k matrix, where m is the number of columns in the counts matrix
X, and k > 1 is the rank of the matrix factorization (equivalently, the number of “topics”). All entries ofFshould be non-negative. WhenFandLare not provided, input argumentkshould be specified instead.- L
An optional argument giving the initial estimate of the loadings (also known as “activations”). It should be an n x k matrix, where n is the number of rows in the counts matrix
X, and k > 1 is the rank of the matrix factorization (equivalently, the number of “topics”). All entries ofLshould be non-negative. WhenFandLare not provided, input argumentkshould be specified instead.- beta
Initial setting of the extrapolation parameter. This is \(beta\) in Algorithm 3 of Ang & Gillis (2019).
- betamax
Initial setting for the upper bound on the extrapolation parameter. This is \(\bar{\gamma}\) in Algorithm 3 of Ang & Gillis (2019).
- clusters
A factor specifying a grouping, or clustering, of the rows of
X.- ...
Additional arguments passed to
init_poisson_nmf.
Value
init_poisson_nmf and fit_poisson_nmf both
return an object capturing the optimization algorithm state (for
init_poisson_nmf, this is the initial state). It is a list
with the following elements:
- F
A matrix containing the current best estimates of the factors.
- L
A matrix containing the current best estimates of the loadings.
- Fn
A matrix containing the non-extrapolated factor estimates. If extrapolation is not used,
FnandFwill be the same.- Ln
A matrix containing the non-extrapolated estimates of the loadings. If extrapolation is not used,
LnandLwill be the same.- Fy
A matrix containing the extrapolated factor estimates. If the extrapolation scheme is not used,
FyandFwill be the same.- Ly
A matrix containing the extrapolated estimates of the loadings. If extrapolation is not used,
LyandLwill be the same.- loss
Value of the objective (“loss”) function computed at the current best estimates of the factors and loadings.
- loss.fnly
Value of the objective (“loss”) function computed at the extrapolated solution for the loadings (
Ly) and the non-extrapolated solution for the factors (Fn). This is used internally to implement the extrapolated updates.- iter
The number of the most recently completed iteration.
- beta
The extrapolation parameter, \(beta\) in Algorithm 3 of Ang & Gillis (2019).
- betamax
Upper bound on the extrapolation parameter. This is \(\bar{\gamma}\) in Algorithm 3 of Ang & Gillis (2019).
- beta0
The setting of the extrapolation parameter at the last iteration that improved the solution.
- progress
A data frame containing detailed information about the algorithm's progress. The data frame should have at most
numiterrows. The columns of the data frame are: “iter”, the iteration number; “loglik”, the Poisson NMF log-likelihood at the current best factor and loading estimates; “loglik.multinom”, the multinomial topic model log-likelihood at the current best factor and loading estimates; “dev”, the deviance at the current best factor and loading estimates; “res”, the maximum residual of the Karush-Kuhn-Tucker (KKT) first-order optimality conditions at the current best factor and loading estimates; “delta.f”, the largest change in the factors matrix; “delta.l”, the largest change in the loadings matrix; “nonzeros.f”, the proportion of entries in the factors matrix that are nonzero; “nonzeros.l”, the proportion of entries in the loadings matrix that are nonzero; “extrapolate”, which is 1 if extrapolation is used, otherwise it is 0; “beta”, the setting of the extrapolation parameter; “betamax”, the setting of the extrapolation parameter upper bound; and “timing”, the elapsed time in seconds (recorded usingproc.time).
Details
In Poisson non-negative matrix factorization (Lee & Seung,
2001), counts \(x_{ij}\) in the \(n \times m\) matrix, \(X\),
are modeled by the Poisson distribution: $$x_{ij} \sim
\mathrm{Poisson}(\lambda_{ij}).$$ Each Poisson rate,
\(\lambda_{ij}\), is a linear combination of parameters
\(f_{jk} \geq 0, l_{ik} \geq 0\) to be fitted to the data:
$$\lambda_{ij} = \sum_{k=1}^K l_{ik} f_{jk},$$ in which \(K\)
is a user-specified tuning parameter specifying the rank of the
matrix factorization. Function fit_poisson_nmf computes
maximum-likelihood estimates (MLEs) of the parameters. For
additional mathematical background, and an explanation of how
Poisson NMF is connected to topic modeling, see the vignette:
vignette(topic = "relationship",package = "fastTopics").
Using this function requires some care; only minimal argument checking is performed, and error messages may not be helpful.
The EM and multiplicative updates are simple and fast, but can be
slow to converge to a stationary point. When control$numiter
= 1, the EM and multiplicative updates are mathematically
equivalent to the multiplicative updates, and therefore share the
same convergence properties. However, the implementation of the EM
updates is quite different; in particular, the EM updates are more
suitable for sparse counts matrices. The implementation of the
multiplicative updates is adapted from the MATLAB code by Daichi
Kitamura http://d-kitamura.net.
Since the multiplicative updates are implemented using standard matrix operations, the speed is heavily dependent on the BLAS/LAPACK numerical libraries used. In particular, using optimized implementations such as OpenBLAS or Intel MKL can result in much improved performance of the multiplcative updates.
The cyclic co-ordinate descent (CCD) and sequential co-ordinate descent (SCD) updates adopt the same optimization strategy, but differ in the implementation details. In practice, we have found that the CCD and SCD updates arrive at the same solution when initialized “sufficiently close” to a stationary point. The CCD implementation is adapted from the C++ code developed by Cho-Jui Hsieh and Inderjit Dhillon, which is available for download at https://www.cs.utexas.edu/~cjhsieh/nmf/. The SCD implementation is based on version 0.4-3 of the ‘NNLM’ package.
An additional re-scaling step is performed after each update to promote numerical stability.
We use three measures of progress for the model fitting: (1)
improvement in the log-likelihood (or deviance), (2) change in the
model parameters, and (3) the residuals of the Karush-Kuhn-Tucker
(KKT) first-order conditions. As the iterates approach a stationary
point of the loss function, the change in the model parameters
should be small, and the residuals of the KKT system should vanish.
Use plot_progress to plot the improvement in the
solution over time.
See fit_topic_model for additional guidance on model
fitting, particularly for large or complex data sets.
The control argument is a list in which any of the
following named components will override the default optimization
algorithm settings (as they are defined by
fit_poisson_nmf_control_default):
numiterNumber of “inner loop” iterations to run when performing and update of the factors or loadings. This must be set to 1 for
method = "mu"andmethod = "ccd".ncNumber of RcppParallel threads to use for the updates. When
ncisNA, the number of threads is determined by callingdefaultNumThreads. This setting is ignored for the multiplicative upates (method = "mu"). Please note that best multithreading performance is typically achieved when the number of BLAS threads is set to 1, but controlling this in R is not always possible; see the “nc.blas” control option for more information.nc.blasNumber of threads used in the numerical linear algebra library (e.g., OpenBLAS), if available. Typically setting this to 1 (i.e., no multithreading) results in best performance. Note that setting the number of BLAS threads relies on the RhpcBLASctl package, and may not work for all multithreaded BLAS libraries.
min.delta.loglikStop performing updates if the difference in the Poisson NMF log-likelihood between two successive updates is less than
min.delta.loglik. This should not be kept at zero whencontrol$extrapolate = TRUEbecause the extrapolated updates are expected to occasionally keep the likelihood unchanged. Ignored ifmin.delta.loglik < 0.min.resStop performing updates if the maximum KKT residual is less than
min.res. Ignored ifmin.res < 0.minvalA small, positive constant used to safeguard the multiplicative updates. The safeguarded updates are implemented as
F <- pmax(F1,minval)andL <- pmax(L1,minval), whereF1andL1are the factors and loadings matrices obtained by applying an update. This is motivated by Theorem 1 of Gillis & Glineur (2012). Settingminval = 0is allowed, but some methods are not guaranteed to converge to a stationary point without this safeguard, and a warning will be given in this case.extrapolateWhen
extrapolate = TRUE, the extrapolation scheme of Ang & Gillis (2019) is used.extrapolate.resetTo promote better numerical stability of the extrapolated updates, they are “reset” every so often. This parameter determines the number of iterations to wait before resetting.
beta.increaseWhen the extrapolated update improves the solution, scale the extrapolation parameter by this amount.
beta.reduceWhen the extrapolaaed update does not improve the solution, scale the extrapolation parameter by this amount.
betamax.increaseWhen the extrapolated update improves the solution, scale the extrapolation parameter by this amount.
epsA small, non-negative number that is added to the terms inside the logarithms to sidestep computing logarithms of zero. This prevents numerical problems at the cost of introducing a small inaccuracy in the solution. Increasing this number may lead to faster convergence but possibly a less accurate solution.
zero.thresholdA small, non-negative number used to determine which entries of the solution are exactly zero. Any entries that are less than or equal to
zero.thresholdare considered to be exactly zero.
An additional setting, control$init.numiter, controls the
number of sequential co-ordinate descent (SCD) updates that are
performed to initialize the loadings matrix when init.method
= "topicscore".
References
Ang, A. and Gillis, N. (2019). Accelerating nonnegative matrix factorization algorithms using extrapolation. Neural Computation 31, 417–439.
Cichocki, A., Cruces, S. and Amari, S. (2011). Generalized alpha-beta divergences and their application to robust nonnegative matrix factorization. Entropy 13, 134–170.
Gillis, N. and Glineur, F. (2012). Accelerated multiplicative
updates and hierarchical ALS algorithms for nonnegative matrix
factorization. Neural Computation 24, 1085–1105.
Hsieh, C.-J. and Dhillon, I. (2011). Fast coordinate descent methods with variable selection for non-negative matrix factorization. In Proceedings of the 17th ACM SIGKDD international conference on Knowledge discovery and data mining, p. 1064-1072
Lee, D. D. and Seung, H. S. (2001). Algorithms for non-negative matrix factorization. In Advances in Neural Information Processing Systems 13, 556–562.
Lin, X. and Boutros, P. C. (2018). Optimization and expansion of non-negative matrix factorization. BMC Bioinformatics 21, 7.
Ke, Z. & Wang, M. (2017). A new SVD approach to optimal topic estimation. arXiv https://arxiv.org/abs/1704.07016
See also
Examples
# Simulate a (sparse) 80 x 100 counts matrix.
library(Matrix)
set.seed(1)
X <- simulate_count_data(80,100,k = 3,sparse = TRUE)$X
# Remove columns (words) that do not appear in any row (document).
X <- X[,colSums(X > 0) > 0]
# Run 10 EM updates to find a good initialization.
fit0 <- fit_poisson_nmf(X,k = 3,numiter = 10,method = "em")
#> Initializing factors using Topic SCORE algorithm.
#> Initializing loadings by running 10 SCD updates.
#> Fitting rank-3 Poisson NMF to 80 x 100 sparse matrix.
#> Running at most 10 EM updates, without extrapolation (fastTopics 0.7-38).
# Fit the Poisson NMF model by running 50 EM updates.
fit_em <- fit_poisson_nmf(X,fit0 = fit0,numiter = 50,method = "em")
#> Fitting rank-3 Poisson NMF to 80 x 100 sparse matrix.
#> Running at most 50 EM updates, without extrapolation (fastTopics 0.7-38).
# Fit the Poisson NMF model by running 50 extrapolated SCD updates.
fit_scd <- fit_poisson_nmf(X,fit0 = fit0,numiter = 50,method = "scd",
control = list(extrapolate = TRUE))
#> Fitting rank-3 Poisson NMF to 80 x 100 sparse matrix.
#> Running at most 50 SCD updates, with extrapolation (fastTopics 0.7-38).
# Compare the two fits.
fits <- list(em = fit_em,scd = fit_scd)
compare_fits(fits)
#> k loglik dev loglik.diff dev.diff res nonzeros.f
#> em 3 -8316.196 7719.679 0.000000 0.000000 1.821374e-01 0.8533333
#> scd 3 -8311.760 7710.809 4.435338 8.870676 1.283011e-05 0.8766667
#> nonzeros.l numiter runtime
#> em 0.8875000 60 0.089
#> scd 0.8166667 60 0.161
plot_progress(fits,y = "loglik")
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the fastTopics package.
#> Please report the issue at
#> <https://github.com/stephenslab/fastTopics/issues>.
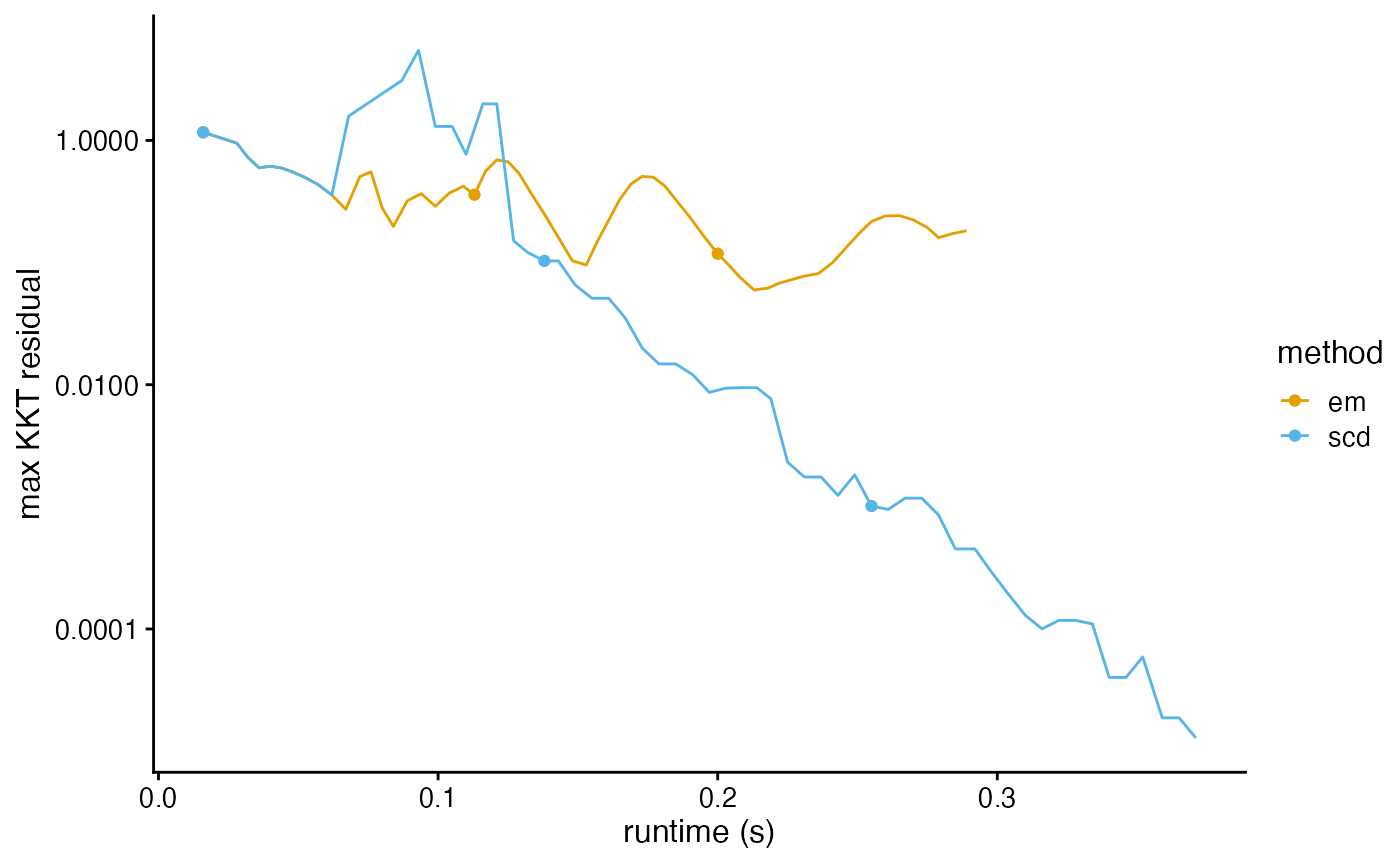
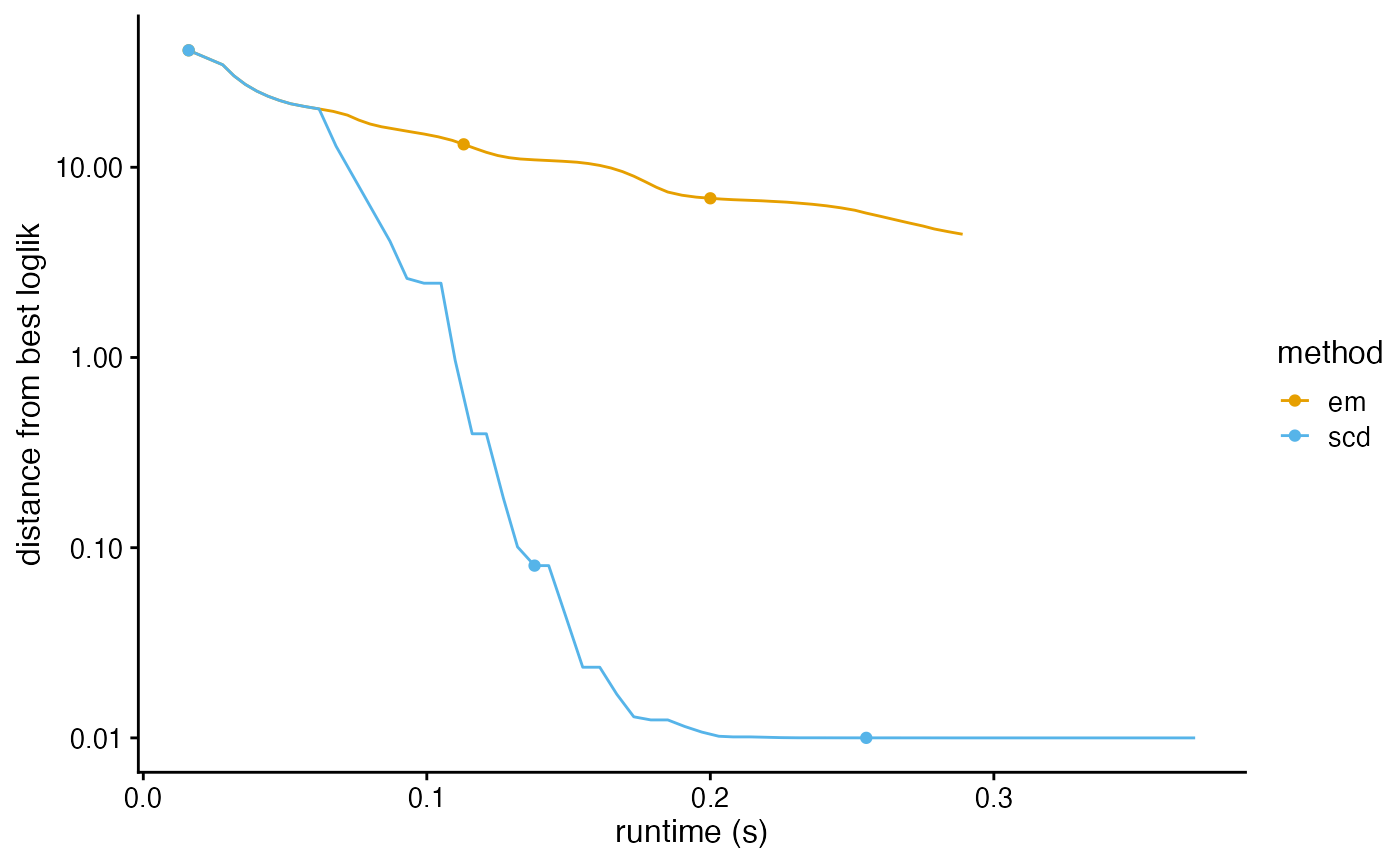 plot_progress(fits,y = "res")
plot_progress(fits,y = "res")
 # Recover the topic model. After this step, the L matrix contains the
# mixture proportions ("loadings"), and the F matrix contains the
# word frequencies ("factors").
fit_multinom <- poisson2multinom(fit_scd)
# Recover the topic model. After this step, the L matrix contains the
# mixture proportions ("loadings"), and the F matrix contains the
# word frequencies ("factors").
fit_multinom <- poisson2multinom(fit_scd)