Low-dimensional Embeddings from Poisson NMF or Multinomial Topic Model
Source:R/embeddings.R
embeddings_from_topics.RdLightweight interface for rapidly producing low-dimensional embeddings from matrix factorizations or multinomial topic models. The defaults used are more suitable for producing embeddings from matrix factorizations or topic models.
pca_from_topics(fit, dims = 2, center = TRUE, scale. = FALSE, ...)
tsne_from_topics(
fit,
dims = 2,
pca = FALSE,
normalize = FALSE,
perplexity = 100,
theta = 0.1,
max_iter = 1000,
eta = 200,
check_duplicates = FALSE,
verbose = TRUE,
...
)
umap_from_topics(
fit,
dims = 2,
n_neighbors = 30,
metric = "euclidean",
scale = "none",
pca = NULL,
verbose = TRUE,
...
)Arguments
- fit
An object of class “poisson_nmf_fit” or “multinom_topic_model_fit”.
- dims
The number of dimensions in the embedding. In
tsne_from_topics, this is passed as argument “dims” toRtsne. Inumap_from_topics, this is passed as argument “n_components” toumap.- center
A logical value indicating whether columns of
fit$Lshould be zero-centered before performing PCA; passed as argument “center” toprcomp.- scale.
A logical value indicating whether columns of
fit$Lshould be scaled to have unit variance prior to performing PCA; passed as argument “scale.” toprcomp.- ...
- pca
Whether to perform a PCA processing step in t-SNE or UMAP; passed as argument “pca” to
Rtsneorumap.- normalize
Whether to normalize the data prior to running t-SNE; passed as argument “normalize” to
Rtsne.- perplexity
t-SNE perplexity parameter, passed as argument “perplexity” to
Rtsne. The perplexity is automatically revised if it is too large; seeRtsnefor more information.- theta
t-SNE speed/accuracy trade-off parameter; passed as argument “theta” to
Rtsne.- max_iter
Maximum number of t-SNE iterations; passed as argument “max_iter” to
Rtsne.- eta
t-SNE learning rate parameter; passed as argument “eta” to
Rtsne.- check_duplicates
When
check_duplicates = TRUE, checks whether there are duplicate rows infit$L; passed as argument “check_duplicates” toRtsne.- verbose
If
verbose = TRUE, progress updates are printed; passed as argument “verbose” toRtsneorumap.- n_neighbors
Number of nearest neighbours in manifold approximation; passed as argument “n_neighbors” to
umap.- metric
Distance matrix used to find nearest neighbors; passed as argument “metric” to
umap.- scale
Scaling to apply to
fit$L; passed as argument “scale” toumap.
Value
An n x d matrix containing the embedding, where n is the
number of rows of fit$L, and d = dims.
Details
Note that since tsne_from_topics and
umap_from_topics use nonlinear transformations of the data,
distances between points are generally less interpretable than a
linear transformation obtained by, say, PCA.
References
Kobak, D. and Berens, P. (2019). The art of using t-SNE for single-cell transcriptomics. Nature Communications 10, 5416. doi:10.1038/s41467-019-13056-x
Examples
library(ggplot2)
library(cowplot)
set.seed(1)
data(pbmc_facs)
# Get the Poisson NMF and multinomial topic model fit to the PBMC data.
fit1 <- multinom2poisson(pbmc_facs$fit)
fit2 <- pbmc_facs$fit
fit2 <- poisson2multinom(fit1)
# Compute the first two PCs of the loadings matrix (for the topic
# model, fit2, the loadings are the topic proportions).
Y1 <- pca_from_topics(fit1)
Y2 <- pca_from_topics(fit2)
subpop <- pbmc_facs$samples$subpop
quickplot(Y1[,1],Y1[,2],color = subpop) + theme_cowplot()
#> Warning: `qplot()` was deprecated in ggplot2 3.4.0.
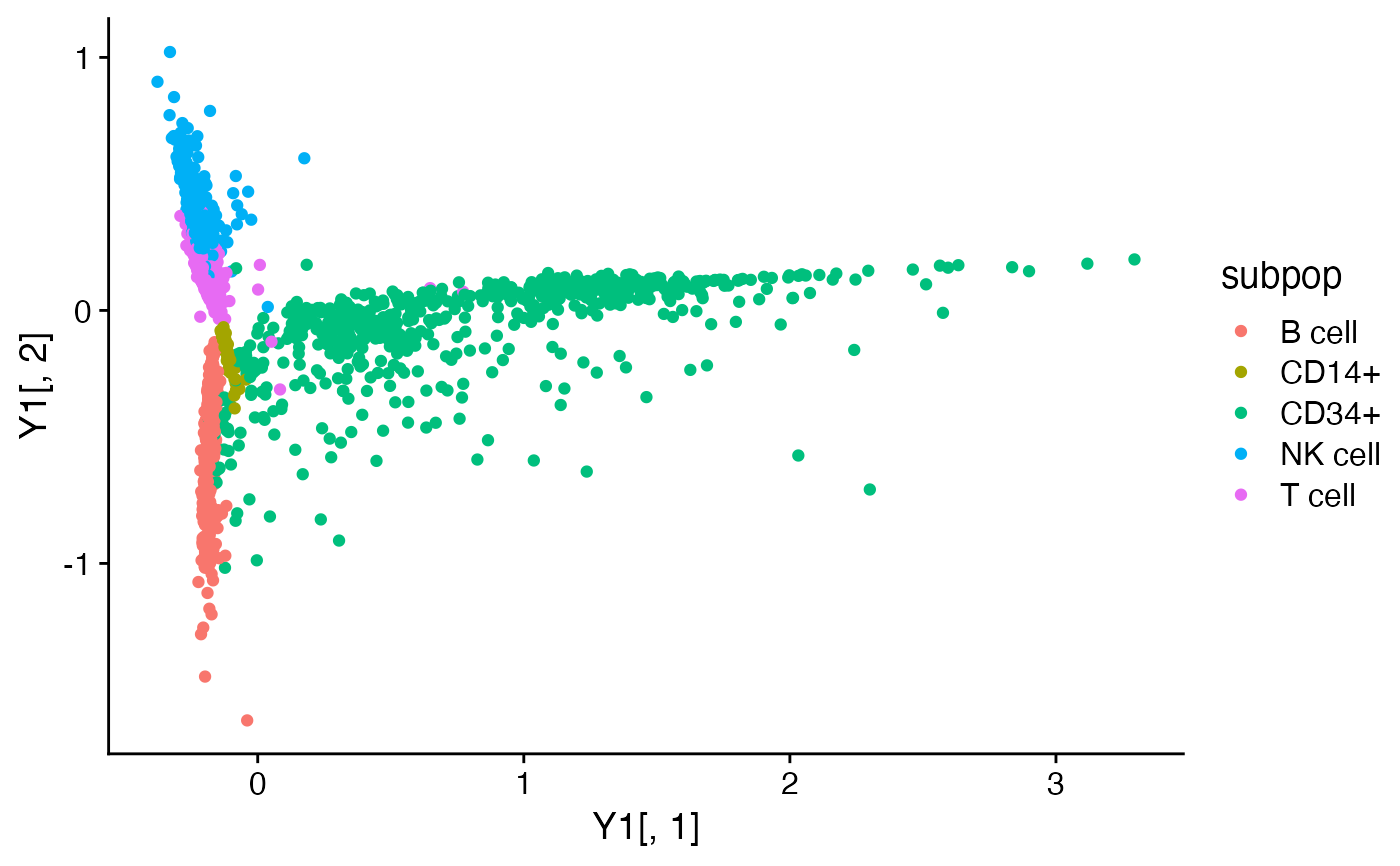 quickplot(Y2[,1],Y2[,2],color = subpop) + theme_cowplot()
quickplot(Y2[,1],Y2[,2],color = subpop) + theme_cowplot()
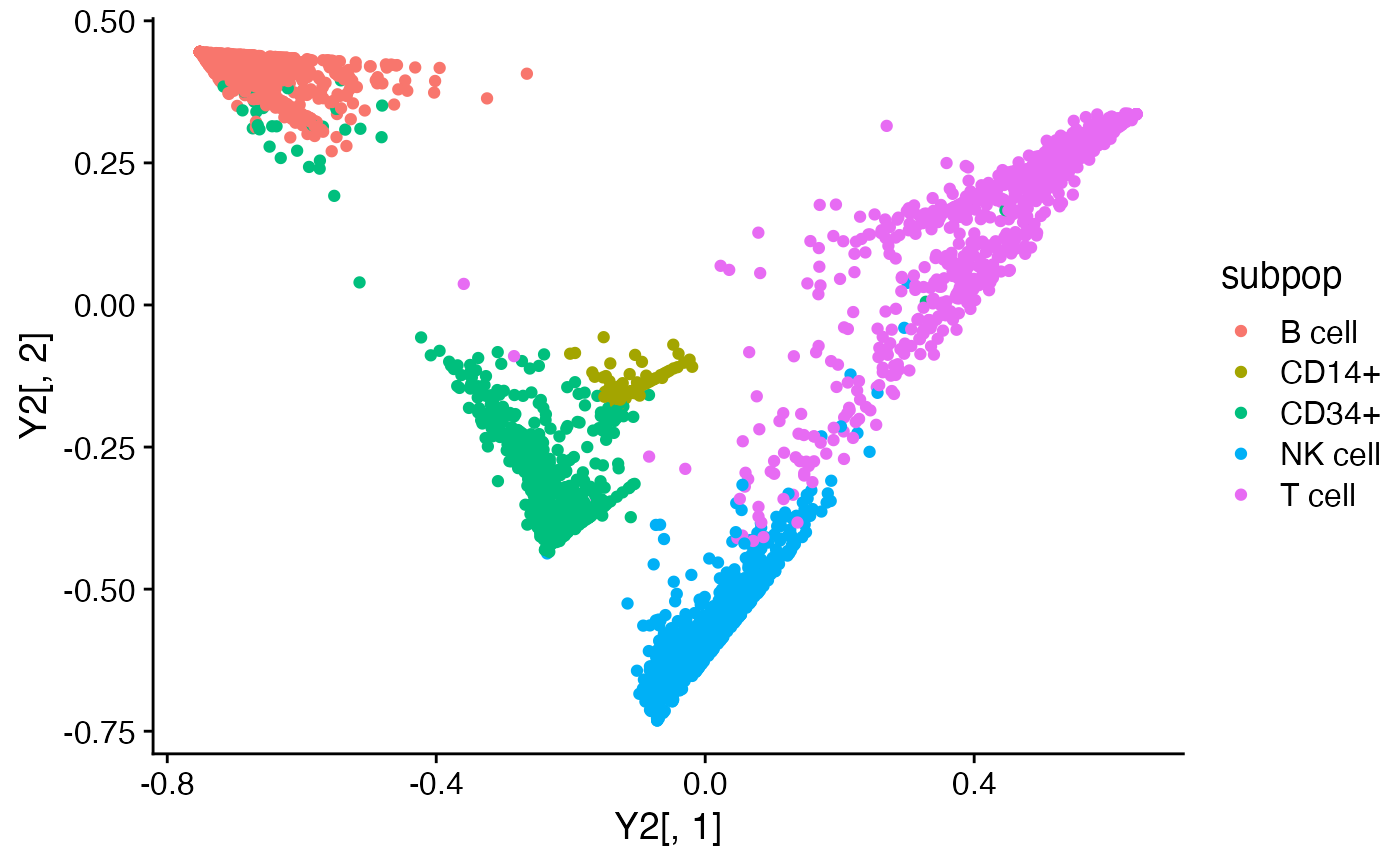 # Compute a 2-d embedding of the loadings using t-SNE.
# \donttest{
Y1 <- tsne_from_topics(fit1)
#> Read the 3774 x 6 data matrix successfully!
#> Using no_dims = 2, perplexity = 100.000000, and theta = 0.100000
#> Computing input similarities...
#> Building tree...
#> Done in 0.55 seconds (sparsity = 0.103577)!
#> Learning embedding...
#> Iteration 50: error is 67.907680 (50 iterations in 1.36 seconds)
#> Iteration 100: error is 55.093036 (50 iterations in 0.95 seconds)
#> Iteration 150: error is 53.522705 (50 iterations in 0.90 seconds)
#> Iteration 200: error is 52.982723 (50 iterations in 0.90 seconds)
#> Iteration 250: error is 52.700813 (50 iterations in 0.90 seconds)
#> Iteration 300: error is 0.869868 (50 iterations in 1.11 seconds)
#> Iteration 350: error is 0.639018 (50 iterations in 1.19 seconds)
#> Iteration 400: error is 0.537143 (50 iterations in 1.17 seconds)
#> Iteration 450: error is 0.482721 (50 iterations in 1.17 seconds)
#> Iteration 500: error is 0.449646 (50 iterations in 1.15 seconds)
#> Iteration 550: error is 0.427580 (50 iterations in 1.14 seconds)
#> Iteration 600: error is 0.412004 (50 iterations in 1.14 seconds)
#> Iteration 650: error is 0.400458 (50 iterations in 1.12 seconds)
#> Iteration 700: error is 0.391642 (50 iterations in 1.13 seconds)
#> Iteration 750: error is 0.384759 (50 iterations in 1.13 seconds)
#> Iteration 800: error is 0.379201 (50 iterations in 1.14 seconds)
#> Iteration 850: error is 0.374679 (50 iterations in 1.14 seconds)
#> Iteration 900: error is 0.370959 (50 iterations in 1.15 seconds)
#> Iteration 950: error is 0.367860 (50 iterations in 1.14 seconds)
#> Iteration 1000: error is 0.365237 (50 iterations in 1.12 seconds)
#> Fitting performed in 22.15 seconds.
Y2 <- tsne_from_topics(fit2)
#> Read the 3774 x 6 data matrix successfully!
#> Using no_dims = 2, perplexity = 100.000000, and theta = 0.100000
#> Computing input similarities...
#> Building tree...
#> Done in 0.65 seconds (sparsity = 0.101133)!
#> Learning embedding...
#> Iteration 50: error is 65.682366 (50 iterations in 1.56 seconds)
#> Iteration 100: error is 53.576821 (50 iterations in 0.97 seconds)
#> Iteration 150: error is 51.735807 (50 iterations in 0.87 seconds)
#> Iteration 200: error is 51.078410 (50 iterations in 0.84 seconds)
#> Iteration 250: error is 50.736216 (50 iterations in 0.83 seconds)
#> Iteration 300: error is 0.804715 (50 iterations in 1.03 seconds)
#> Iteration 350: error is 0.580748 (50 iterations in 1.08 seconds)
#> Iteration 400: error is 0.481837 (50 iterations in 1.07 seconds)
#> Iteration 450: error is 0.429849 (50 iterations in 1.06 seconds)
#> Iteration 500: error is 0.398384 (50 iterations in 1.06 seconds)
#> Iteration 550: error is 0.377602 (50 iterations in 1.05 seconds)
#> Iteration 600: error is 0.362985 (50 iterations in 1.05 seconds)
#> Iteration 650: error is 0.352226 (50 iterations in 1.05 seconds)
#> Iteration 700: error is 0.344063 (50 iterations in 1.03 seconds)
#> Iteration 750: error is 0.337688 (50 iterations in 1.02 seconds)
#> Iteration 800: error is 0.332612 (50 iterations in 1.02 seconds)
#> Iteration 850: error is 0.328442 (50 iterations in 1.01 seconds)
#> Iteration 900: error is 0.325026 (50 iterations in 1.02 seconds)
#> Iteration 950: error is 0.322085 (50 iterations in 1.00 seconds)
#> Iteration 1000: error is 0.319664 (50 iterations in 1.00 seconds)
#> Fitting performed in 20.62 seconds.
quickplot(Y1[,1],Y1[,2],color = subpop) + theme_cowplot()
# Compute a 2-d embedding of the loadings using t-SNE.
# \donttest{
Y1 <- tsne_from_topics(fit1)
#> Read the 3774 x 6 data matrix successfully!
#> Using no_dims = 2, perplexity = 100.000000, and theta = 0.100000
#> Computing input similarities...
#> Building tree...
#> Done in 0.55 seconds (sparsity = 0.103577)!
#> Learning embedding...
#> Iteration 50: error is 67.907680 (50 iterations in 1.36 seconds)
#> Iteration 100: error is 55.093036 (50 iterations in 0.95 seconds)
#> Iteration 150: error is 53.522705 (50 iterations in 0.90 seconds)
#> Iteration 200: error is 52.982723 (50 iterations in 0.90 seconds)
#> Iteration 250: error is 52.700813 (50 iterations in 0.90 seconds)
#> Iteration 300: error is 0.869868 (50 iterations in 1.11 seconds)
#> Iteration 350: error is 0.639018 (50 iterations in 1.19 seconds)
#> Iteration 400: error is 0.537143 (50 iterations in 1.17 seconds)
#> Iteration 450: error is 0.482721 (50 iterations in 1.17 seconds)
#> Iteration 500: error is 0.449646 (50 iterations in 1.15 seconds)
#> Iteration 550: error is 0.427580 (50 iterations in 1.14 seconds)
#> Iteration 600: error is 0.412004 (50 iterations in 1.14 seconds)
#> Iteration 650: error is 0.400458 (50 iterations in 1.12 seconds)
#> Iteration 700: error is 0.391642 (50 iterations in 1.13 seconds)
#> Iteration 750: error is 0.384759 (50 iterations in 1.13 seconds)
#> Iteration 800: error is 0.379201 (50 iterations in 1.14 seconds)
#> Iteration 850: error is 0.374679 (50 iterations in 1.14 seconds)
#> Iteration 900: error is 0.370959 (50 iterations in 1.15 seconds)
#> Iteration 950: error is 0.367860 (50 iterations in 1.14 seconds)
#> Iteration 1000: error is 0.365237 (50 iterations in 1.12 seconds)
#> Fitting performed in 22.15 seconds.
Y2 <- tsne_from_topics(fit2)
#> Read the 3774 x 6 data matrix successfully!
#> Using no_dims = 2, perplexity = 100.000000, and theta = 0.100000
#> Computing input similarities...
#> Building tree...
#> Done in 0.65 seconds (sparsity = 0.101133)!
#> Learning embedding...
#> Iteration 50: error is 65.682366 (50 iterations in 1.56 seconds)
#> Iteration 100: error is 53.576821 (50 iterations in 0.97 seconds)
#> Iteration 150: error is 51.735807 (50 iterations in 0.87 seconds)
#> Iteration 200: error is 51.078410 (50 iterations in 0.84 seconds)
#> Iteration 250: error is 50.736216 (50 iterations in 0.83 seconds)
#> Iteration 300: error is 0.804715 (50 iterations in 1.03 seconds)
#> Iteration 350: error is 0.580748 (50 iterations in 1.08 seconds)
#> Iteration 400: error is 0.481837 (50 iterations in 1.07 seconds)
#> Iteration 450: error is 0.429849 (50 iterations in 1.06 seconds)
#> Iteration 500: error is 0.398384 (50 iterations in 1.06 seconds)
#> Iteration 550: error is 0.377602 (50 iterations in 1.05 seconds)
#> Iteration 600: error is 0.362985 (50 iterations in 1.05 seconds)
#> Iteration 650: error is 0.352226 (50 iterations in 1.05 seconds)
#> Iteration 700: error is 0.344063 (50 iterations in 1.03 seconds)
#> Iteration 750: error is 0.337688 (50 iterations in 1.02 seconds)
#> Iteration 800: error is 0.332612 (50 iterations in 1.02 seconds)
#> Iteration 850: error is 0.328442 (50 iterations in 1.01 seconds)
#> Iteration 900: error is 0.325026 (50 iterations in 1.02 seconds)
#> Iteration 950: error is 0.322085 (50 iterations in 1.00 seconds)
#> Iteration 1000: error is 0.319664 (50 iterations in 1.00 seconds)
#> Fitting performed in 20.62 seconds.
quickplot(Y1[,1],Y1[,2],color = subpop) + theme_cowplot()
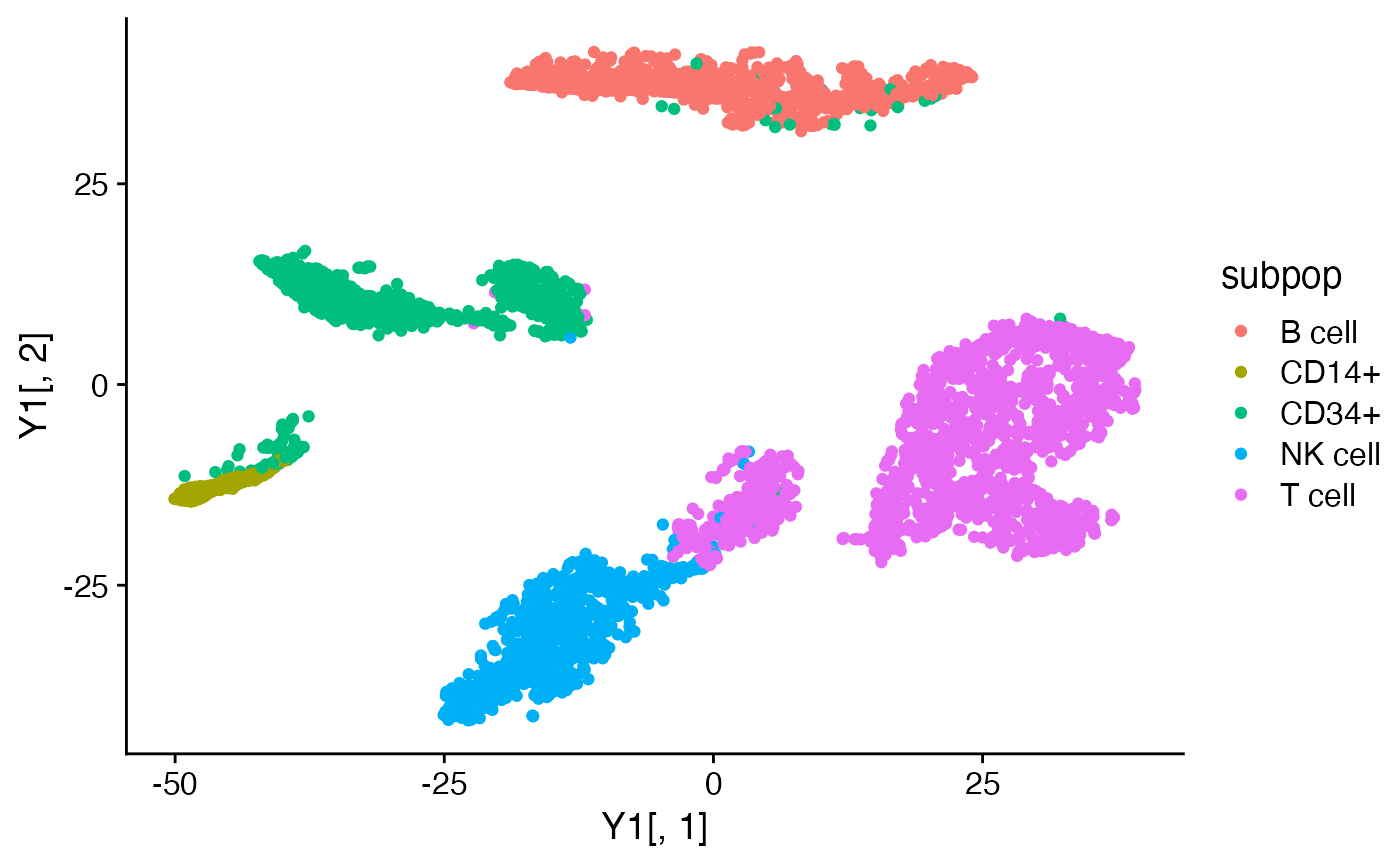 quickplot(Y2[,1],Y2[,2],color = subpop) + theme_cowplot()
quickplot(Y2[,1],Y2[,2],color = subpop) + theme_cowplot()
 # }
# Compute a 2-d embedding of the loadings using UMAP.
# \donttest{
Y1 <- umap_from_topics(fit1)
#> 09:40:22 UMAP embedding parameters a = 1.896 b = 0.8006
#> 09:40:22 Read 3774 rows and found 6 numeric columns
#> 09:40:22 Using FNN for neighbor search, n_neighbors = 30
#> 09:40:22 Commencing smooth kNN distance calibration using 4 threads
#> with target n_neighbors = 30
#> 09:40:23 Initializing from normalized Laplacian + noise (using irlba)
#> 09:40:23 Commencing optimization for 500 epochs, with 139662 positive edges
#> 09:40:23 Using rng type: pcg
#> 09:40:25 Optimization finished
Y2 <- umap_from_topics(fit2)
#> 09:40:25 UMAP embedding parameters a = 1.896 b = 0.8006
#> 09:40:25 Read 3774 rows and found 6 numeric columns
#> 09:40:25 Using FNN for neighbor search, n_neighbors = 30
#> 09:40:25 Commencing smooth kNN distance calibration using 4 threads
#> with target n_neighbors = 30
#> 09:40:25 110 smooth knn distance failures
#> 09:40:26 Initializing from normalized Laplacian + noise (using irlba)
#> 09:40:26 Commencing optimization for 500 epochs, with 137990 positive edges
#> 09:40:26 Using rng type: pcg
#> 09:40:28 Optimization finished
quickplot(Y1[,1],Y1[,2],color = subpop) + theme_cowplot()
# }
# Compute a 2-d embedding of the loadings using UMAP.
# \donttest{
Y1 <- umap_from_topics(fit1)
#> 09:40:22 UMAP embedding parameters a = 1.896 b = 0.8006
#> 09:40:22 Read 3774 rows and found 6 numeric columns
#> 09:40:22 Using FNN for neighbor search, n_neighbors = 30
#> 09:40:22 Commencing smooth kNN distance calibration using 4 threads
#> with target n_neighbors = 30
#> 09:40:23 Initializing from normalized Laplacian + noise (using irlba)
#> 09:40:23 Commencing optimization for 500 epochs, with 139662 positive edges
#> 09:40:23 Using rng type: pcg
#> 09:40:25 Optimization finished
Y2 <- umap_from_topics(fit2)
#> 09:40:25 UMAP embedding parameters a = 1.896 b = 0.8006
#> 09:40:25 Read 3774 rows and found 6 numeric columns
#> 09:40:25 Using FNN for neighbor search, n_neighbors = 30
#> 09:40:25 Commencing smooth kNN distance calibration using 4 threads
#> with target n_neighbors = 30
#> 09:40:25 110 smooth knn distance failures
#> 09:40:26 Initializing from normalized Laplacian + noise (using irlba)
#> 09:40:26 Commencing optimization for 500 epochs, with 137990 positive edges
#> 09:40:26 Using rng type: pcg
#> 09:40:28 Optimization finished
quickplot(Y1[,1],Y1[,2],color = subpop) + theme_cowplot()
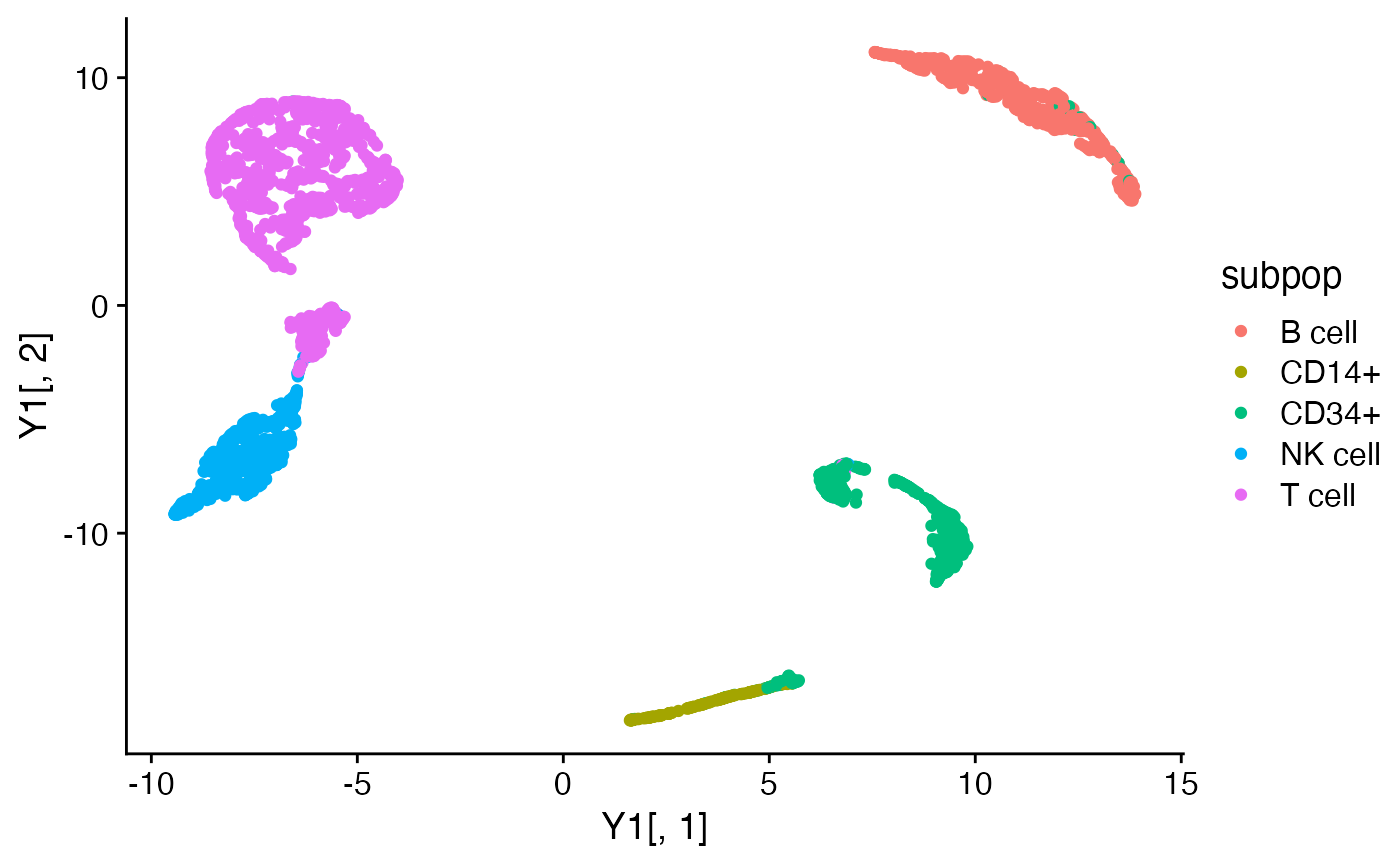 quickplot(Y2[,1],Y2[,2],color = subpop) + theme_cowplot()
quickplot(Y2[,1],Y2[,2],color = subpop) + theme_cowplot()
 # }
# }