Implements methods for differential expression analysis using a topic model. These methods are motivated by gene expression studies, but could have other uses, such as identifying “key words” for topics.
Arguments
- fit
An object of class “poisson_nmf_fit” or “multinom_topic_model_fit”, or an n x k matrix of topic proportions, where k is the number of topics. (The elements in each row of this matrix should sum to 1.) If a Poisson NMF fit is provided as input, the corresponding multinomial topic model fit is automatically recovered using
poisson2multinom.- X
The n x m counts matrix. It can be a sparse matrix (class
"dgCMatrix") or dense matrix (class"matrix").- s
A numeric vector of length n determining how the rates are scaled in the Poisson models. See “Details” for guidance on the choice of
s.- pseudocount
Observations with this value are added to the counts matrix to stabilize maximum-likelihood estimation.
- fit.method
Method used to fit the Poisson models. Note that
fit.method = "glm"is the slowest method, and is mainly used for testing.- shrink.method
Method used to stabilize the posterior mean LFC estimates. When
shrink.method = "ash", the "adaptive shrinkage" method implemented in the ‘ashr’ package is used to compute posterior. Whenshrink.method = "none", no stabilization is performed, and the “raw” posterior mean LFC estimates are returned.- lfc.stat
The log-fold change statistics returned:
lfc.stat = "vsnull", the log-fold change relative to the null;lfc.stat = "le", the “least extreme” LFC; or a topic name or number, in which case the LFC is defined relative to the selected topic. See “Details” for more detailed explanations of these choices.- control
A list of parameters controlling behaviour of the optimization and Monte Carlo algorithms. See ‘Details’.
- verbose
When
verbose = TRUE, progress information is printed to the console.- ...
When
shrink.method = "ash", these are additional arguments passed toash.
Value
A list with the following elements:
- est
The log-fold change maximum-likelihood estimates.
- postmean
Posterior mean LFC estimates.
- lower
Lower limits of estimated HPD intervals. Note that these are not updated by the shrinkage step.
- upper
Upper limits of estimated HPD intervals. Note that these are not updated by the shrinkage step.
- z
z-scores for posterior mean LFC estimates.
- lpval
-log10 two-tailed p-values obtained from the z-scores. When
shrink.method = "ash", this isNA, and the s-values are returned instead (see below).- svalue
s-values returned by
ash. s-values are analogous to q-values, but based on the local false sign rate; see Stephens (2016).- lfsr
When
shrink.method = "ash"only, this output contains the estimated local false sign rates.- ash
When
shrink.method = "ash"only, this output contains theashreturn value (after removing the"data","result"and"call"list elements).- F
Maximum-likelihood estimates of the Poisson model parameters.
- f0
Maximum-likelihood estimates of the null model parameters.
- ar
A vector containing the Metropolis acceptance ratios from each MCMC run.
Details
The methods are based on the Poisson model
$$x_i ~ Poisson(\lambda_i),$$ in which the Poisson rates are
$$\lambda_i = \sum_{j=1}^k s_i l_{ij} f_j,$$ the \(l_{ik}\)
are the topic proportions and the \(f_j\) are the unknowns to be
estimated. This model is applied separately to each column of
X. When \(s_i\) (specified by input argument s) is
equal the total count in row i (this is the default), the Poisson
model will closely approximate a binomial model of the count data,
and the unknowns \(f_j\) will approximate binomial
probabilities. (The Poisson approximation to the binomial is most
accurate when the total counts rowSums(X) are large and the
unknowns \(f_j\) are small.) Other choices for s are
possible, and implement different normalization schemes.
To allow for some flexibility, de_analysis allows for the
log-fold change to be measured in several ways.
One option is to compare against the probability under the null
model: \(LFC(j) = log2(f_j/f_0)\), where \(f_0\) is the single
parameter in the Poisson model \(x_i ~ Poisson(\lambda_i)\) with
rates \(\lambda_i = s_i f_0\). This LFC definition is chosen with
lfc.stat = "vsnull".
Another option is to compare against a chosen topic, k: \(LFC(j)
= log2(f_j/f_k)\). By definition, \(LFC(k)\) is zero, and
statistics such as z-scores and p-values for topic k are set to
NA. This LFC definition is selected by setting
lfc.stat = k.
A final option (which is the default) computes the “least
extreme” LFC, defined as \(LFC(j) = log2(f_j/f_k)\) such that
\(k\) is the topic other than \(j\) that gives the ratio
\(f_j/f_k\) closest to 1. This option is chosen with
lfc.stat = "le".
We recommend setting shrink.method = "ash", which uses the
“adaptive shrinkage” method (Stephens, 2016) to improve
accuracy of the posterior mean estimates and z-scores. We follow
the settings used in lfcShrink from the ‘DESeq2’
package, with type = "ashr".
Note that all LFC statistics are defined using the base-2 logarithm following the conventioned used in differential expression analysis.
The control argument is a list in which any of the
following named components will override the default optimization
algorithm settings (as they are defined by
de_analysis_control_default):
numiterMaximum number of iterations performed in fitting the Poisson models. When
fit.method = "glm", this is passed as argumentmaxitto theglmfunction.minvalA small, positive number. All topic proportions less than this value and greater than
1 - minvalare set to this value.tolControls the convergence tolerance for fitting the Poisson models. When
fit.method = "glm", this is passed as argumentepsilonto functionglm.conf.levelThe size of the highest posterior density (HPD) intervals. Should be a number greater than 0 and less than 1.
nsNumber of Monte Carlo samples simulated by random-walk MCMC for estimating posterior LFC quantities.
rwThe standard deviation of the normal density used to propose new states in the random-walk MCMC.
epsA small, non-negative number added to the terms inside the logarithms to avoid computing logarithms of zero.
ncNumber of threads used in the multithreaded computations. This controls both (a) the number of RcppParallel threads used to fit the factors in the Poisson models, and (b) the number of cores used in
mclapplyfor the MCMC simulation step. Note that mclapply relies on forking hence is not available on Windows; will return an error on Windows unlessnc = 1. Also note that settingnc > 1copies the contents of memorynctimes, which can lead to poor performance if the total resident memory required exceeds available physical memory.nc.blasNumber of threads used in the numerical linear algebra library (e.g., OpenBLAS), if available. For best performance, we recommend setting this to 1 (i.e., no multithreading).
nsplitThe number of data splits used in the multithreaded computations (only relevant when
nc > 1). More splits increase the granularity of the progress bar, but can also slow down the mutithreaded computations by introducing more overhead in the call topblapply.
References
Stephens, M. (2016). False discovery rates: a new deal. Biostatistics 18(2), kxw041. doi:10.1093/biostatistics/kxw041
Zhu, A., Ibrahim, J. G. and Love, M. I. (2019). Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35(12), 2084–2092.
Examples
# Perform a differential expression (DE) analysis using the previously
# fitted multinomial topic model. Note that the de_analysis call could
# take several minutes to complete.
# \donttest{
set.seed(1)
data(pbmc_facs)
de <- de_analysis(pbmc_facs$fit,pbmc_facs$counts)
#> Fitting 16791 Poisson models with k=6 using method="scd".
#> Computing log-fold change statistics from 16791 Poisson models with k=6.
#> Stabilizing posterior log-fold change estimates using adaptive shrinkage.
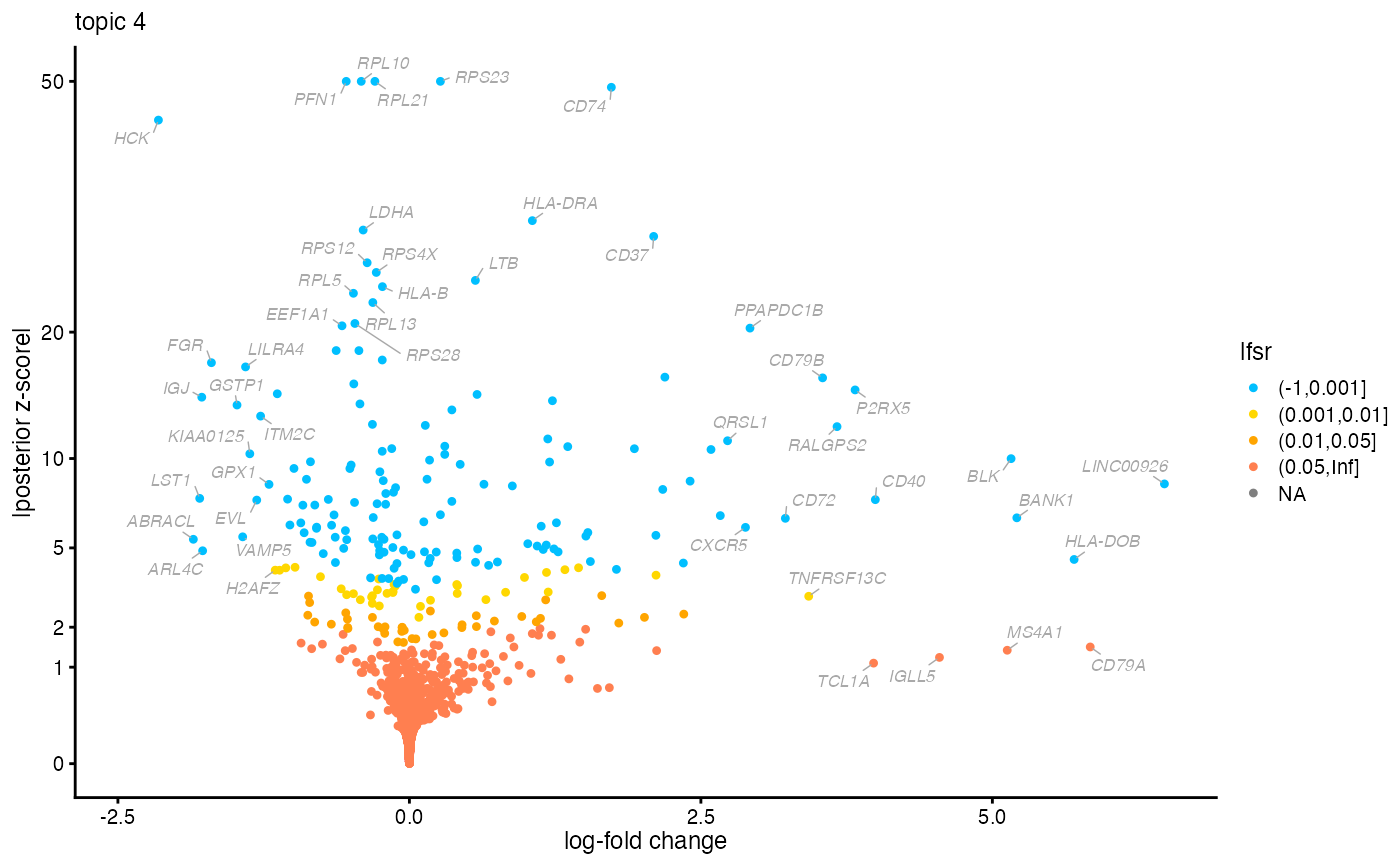
# Compile the DE analysis results for topic 4 into a table, and
# rank the genes by the posterior mean log-fold change, limiting to
# DE genes identified with low lfsr ("local false sign rate").
k <- 4
dat <- data.frame(postmean = de$postmean[,k],
z = de$z[,k],
lfsr = de$lfsr[,k])
rownames(dat) <- with(pbmc_facs$genes,paste(symbol,ensembl,sep = "_"))
dat <- subset(dat,lfsr < 0.01)
dat <- dat[order(dat$postmean,decreasing = TRUE),]
# Genes at the top of this ranking are genes that are much more
# highly expressed in the topic compared to other topics.
head(dat,n = 10)
#> postmean z lfsr
#> LINC00926_ENSG00000247982 6.475599 8.405566 7.976606e-15
#> HLA-DOB_ENSG00000241106 5.700977 4.478842 2.616893e-04
#> BANK1_ENSG00000153064 5.209646 6.491396 6.918661e-09
#> BLK_ENSG00000136573 5.160458 9.992159 2.424077e-21
#> CD40_ENSG00000101017 3.996155 7.485618 9.636518e-12
#> P2RX5_ENSG00000083454 3.822560 15.001932 4.222700e-48
#> RALGPS2_ENSG00000116191 3.667691 12.199455 1.110458e-31
#> CD79B_ENSG00000007312 3.543927 15.986085 7.436217e-55
#> TNFRSF13C_ENSG00000159958 3.424878 3.009488 8.030873e-03
#> CD72_ENSG00000137101 3.224403 6.464962 2.381729e-09
# The genes at the bottom of the ranking are genes that are much less
# expressed in the topic.
tail(dat,n = 10)
#> postmean z lfsr
#> KIAA0125_ENSG00000226777 -1.368692 -10.312766 1.110223e-16
#> LILRA4_ENSG00000239961 -1.405395 -16.906653 1.110223e-16
#> VAMP5_ENSG00000168899 -1.429959 -5.526587 3.007171e-06
#> GSTP1_ENSG00000084207 -1.478443 -13.814083 0.000000e+00
#> FGR_ENSG00000000938 -1.697954 -17.267656 0.000000e+00
#> ARL4C_ENSG00000188042 -1.773366 -4.868172 3.510890e-05
#> IGJ_ENSG00000132465 -1.780658 -14.423687 0.000000e+00
#> LST1_ENSG00000204482 -1.798490 -7.558970 2.199685e-12
#> ABRACL_ENSG00000146386 -1.853036 -5.407003 1.858173e-06
#> HCK_ENSG00000101336 -2.152558 -44.483701 2.220446e-16
# Create a volcano plot from the DE results for topic 4.
volcano_plot(de,k = k,ymax = 50,labels = pbmc_facs$genes$symbol)
 # }
# }