Summary of benchmark on GTEx RNA-seq gene expression data
Wei Wang
2017-08-15
Last updated: 2017-08-24
Code version: 6089d1d
P = 300
#dsc-query omega -o gtexlung300loglik.ipynb --title "compare methods in lung data loglik" --target normalize_data.P normalize_data.n loglik.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash --condition "normalize_data.P = 300" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung300loglik.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash")
score = "loglik_score"
title = "loglik P=300"
p11 = box_per_data(data, methods, score,title)#dsc-query omega -o gtexlung300mse.ipynb --title "compare methods in lung data mse" --target normalize_data.P normalize_data.n mse.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash --condition "normalize_data.P = 300" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung300mse.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash")
score = "mse_score"
title = "MSE P=300"
p12 = box_per_data(data, methods, score,title)#dsc-query omega -o gtexlung300hyvarinen.ipynb --title "compare methods in lung data hyvarinen" --target normalize_data.P normalize_data.n hyvarinen.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash --condition "normalize_data.P = 300" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung300hyvarinen.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash")
score = "hyvarinen_score"
title = "hyvarinen P=300"
p13 = box_per_data(data, methods, score,title)library(cowplot)
plot_grid(p11,p12,p13,ncol = 2)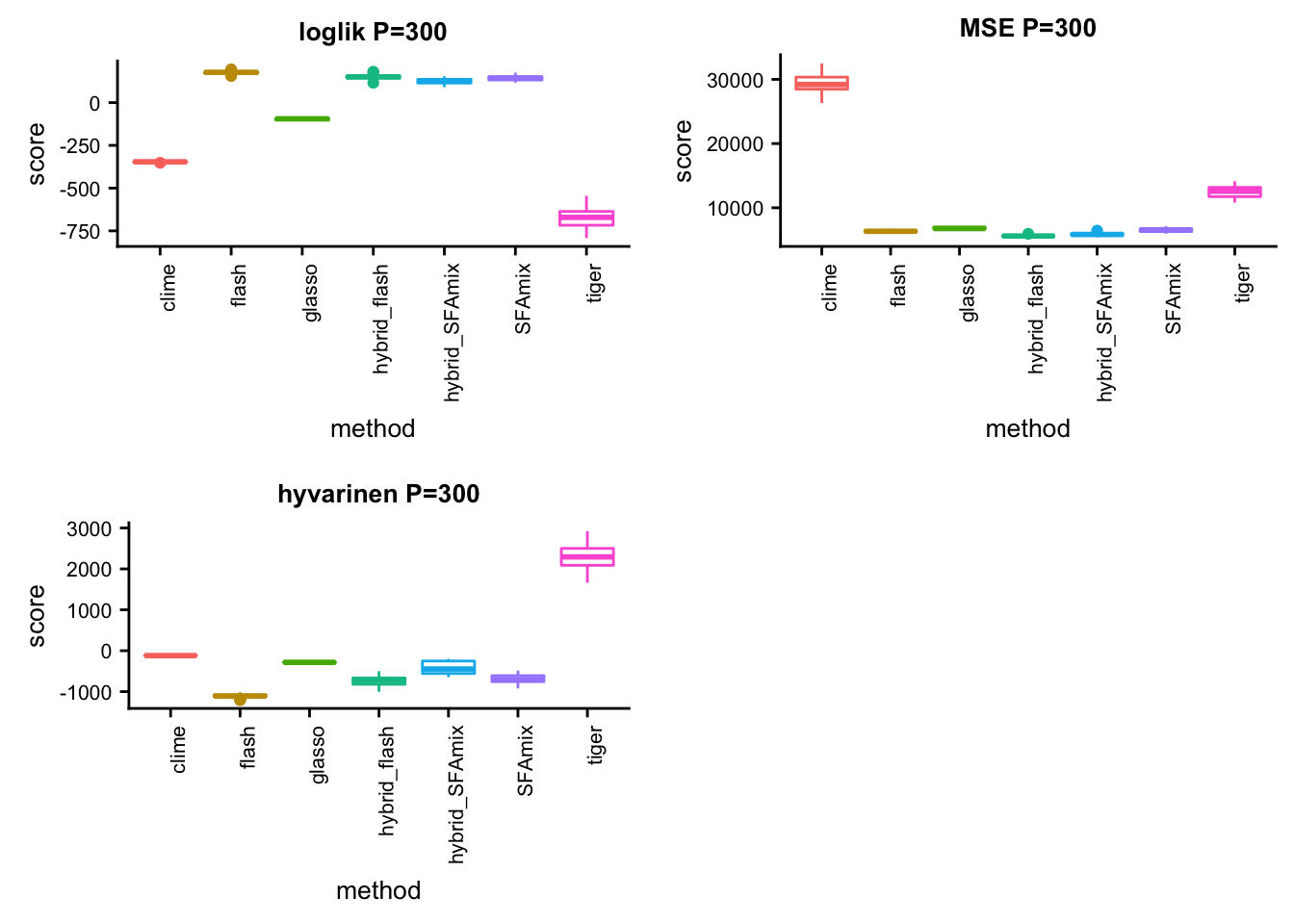
P = 1000
#dsc-query omega -o gtexlung1000loglik.ipynb --title "compare methods in lung data loglik" --target normalize_data.P normalize_data.n loglik.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash --condition "normalize_data.P = 1000" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung1000loglik.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash")
score = "loglik_score"
title = "loglik P=1000"
p21 = box_per_data(data, methods, score,title)#dsc-query omega -o gtexlung1000mse.ipynb --title "compare methods in lung data mse" --target normalize_data.P normalize_data.n mse.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash --condition "normalize_data.P = 1000" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung1000mse.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash")
score = "mse_score"
title = "MSE P=1000"
p22 = box_per_data(data, methods, score,title)#dsc-query omega -o gtexlung1000hyvarinen.ipynb --title "compare methods in lung data hyvarinen" --target normalize_data.P normalize_data.n hyvarinen.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash --condition "normalize_data.P = 1000" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung1000hyvarinen.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash")
score = "hyvarinen_score"
title = "hyvarinen P=1000"
p23 = box_per_data(data, methods, score,title)library(cowplot)
plot_grid(p21,p22,p23,ncol = 2)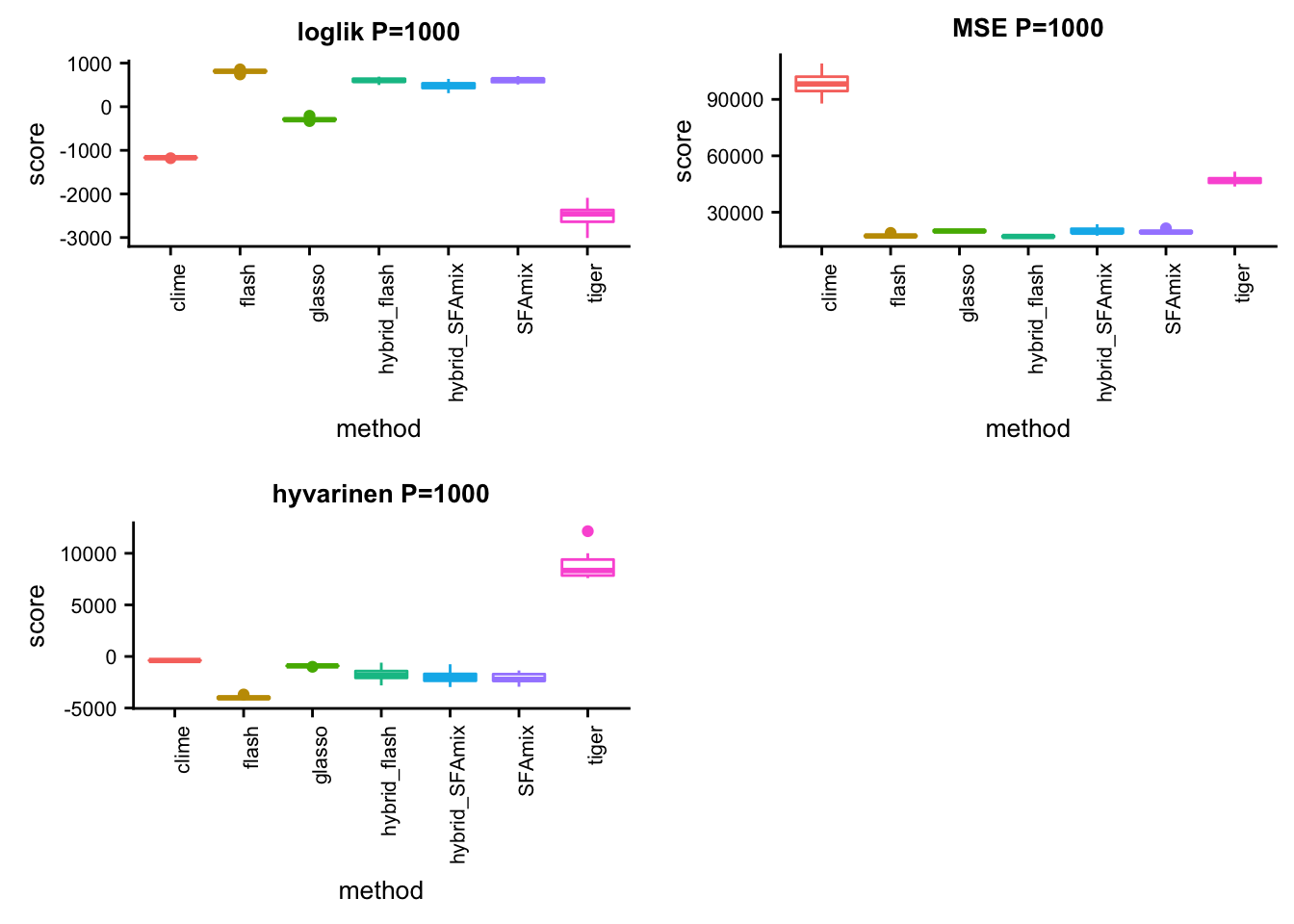
P = 100
#dsc-query omega -o gtexlung100loglik.ipynb --title "compare methods in lung data loglik" --target normalize_data.P normalize_data.n loglik.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash --condition "normalize_data.P = 100" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung100loglik.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash")
score = "loglik_score"
title = "loglik P=100"
p31 = box_per_data(data, methods, score,title)#dsc-query omega -o gtexlung100mse.ipynb --title "compare methods in lung data mse" --target normalize_data.P normalize_data.n mse.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash --condition "normalize_data.P = 100" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung100mse.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash")
score = "mse_score"
title = "MSE P=100"
p32 = box_per_data(data, methods, score,title)#dsc-query omega -o gtexlung100hyvarinen.ipynb --title "compare methods in lung data hyvarinen" --target normalize_data.P normalize_data.n hyvarinen.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash --condition "normalize_data.P = 100" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung100hyvarinen.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash")
score = "hyvarinen_score"
title = "hyvarinen P=100"
p33 = box_per_data(data, methods, score,title)library(cowplot)
plot_grid(p31,p32,p33,ncol = 2)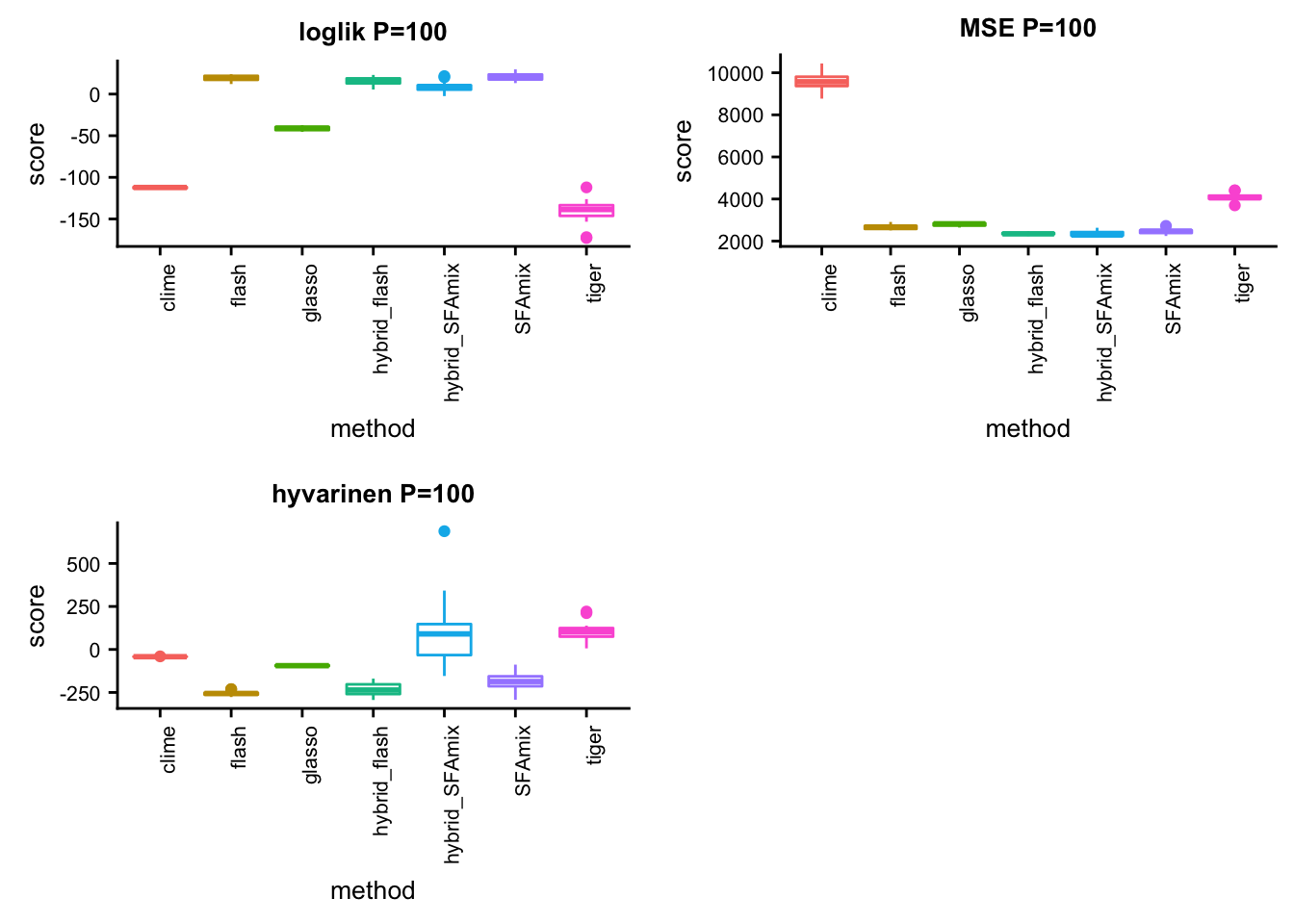
P = 10
#dsc-query omega -o gtexlung10loglik.ipynb --title "compare methods in lung data loglik" --target normalize_data.P normalize_data.n loglik.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash inv_cov --condition "normalize_data.P = 10" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung10loglik.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash","inv_cov")
score = "loglik_score"
title = "loglik P=10"
p41 = box_per_data(data, methods, score,title)#dsc-query omega -o gtexlung10mse.ipynb --title "compare methods in lung data mse" --target normalize_data.P normalize_data.n mse.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash inv_cov --condition "normalize_data.P = 10" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung10mse.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash","inv_cov")
score = "mse_score"
title = "MSE P=10"
p42 = box_per_data(data, methods, score,title)#dsc-query omega -o gtexlung10hyvarinen.ipynb --title "compare methods in lung data hyvarinen" --target normalize_data.P normalize_data.n hyvarinen.score flash SFAmix tiger glasso clime hybrid_SFAmix hybrid_flash inv_cov --condition "normalize_data.P = 10" "normalize_data.n = 200" --language Rsource("../testdata/plotfunction.R")
data = readRDS("../testdata/omega0815/gtexlung10hyvarinen.rds")
methods = c("flash", "SFAmix", "tiger", "glasso", "clime", "hybrid_SFAmix", "hybrid_flash","inv_cov")
score = "hyvarinen_score"
title = "hyvarinen P=10"
p43 = box_per_data(data, methods, score,title)library(cowplot)
plot_grid(p41,p42,p43,ncol = 2)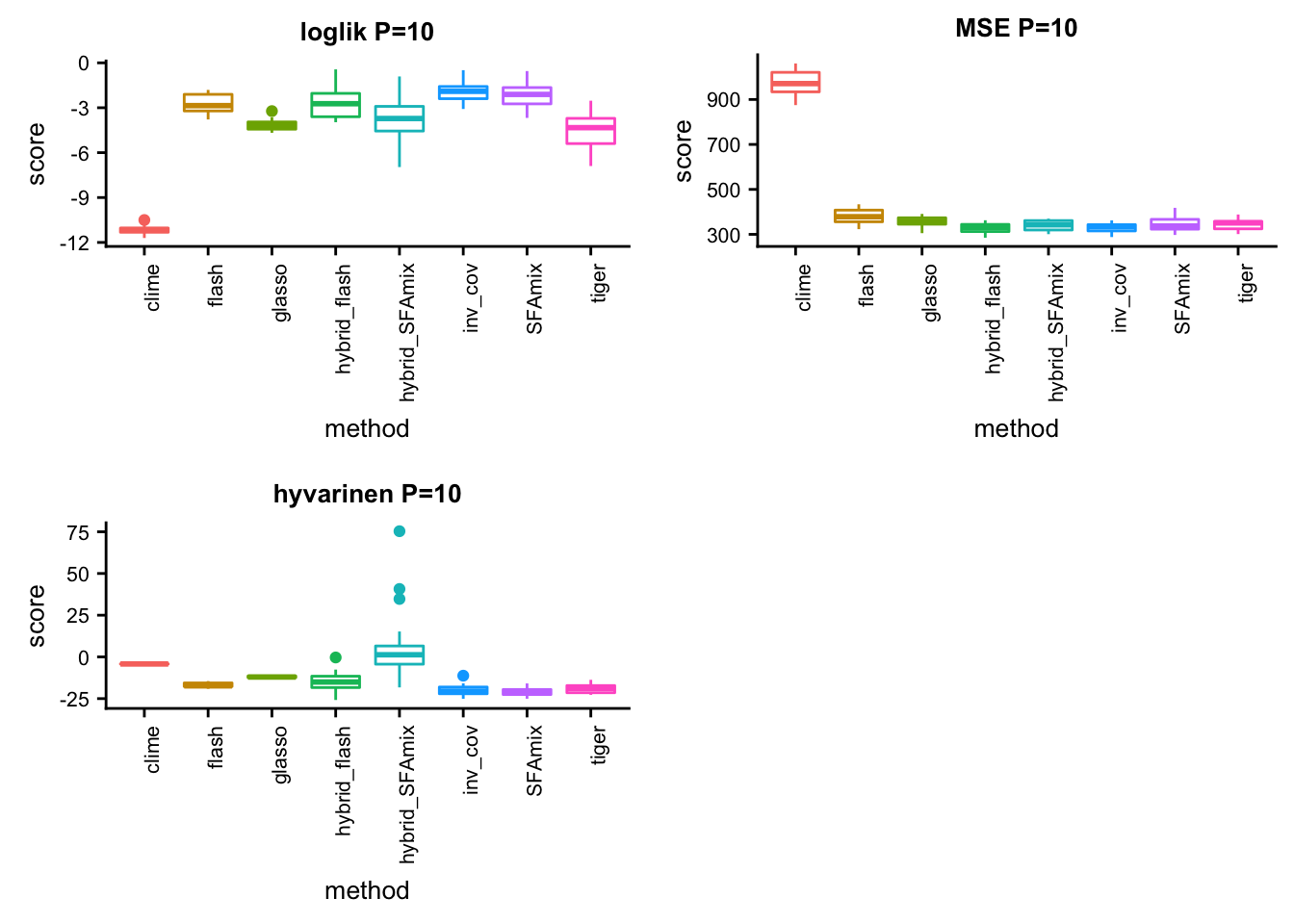
Session information
sessionInfo()R version 3.3.0 (2016-05-03)
Platform: x86_64-apple-darwin13.4.0 (64-bit)
Running under: OS X 10.12.6 (unknown)
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_0.8.0 ggplot2_2.2.1 workflowr_0.4.0 rmarkdown_1.6
loaded via a namespace (and not attached):
[1] Rcpp_0.12.12 digest_0.6.12 rprojroot_1.2 plyr_1.8.4
[5] grid_3.3.0 gtable_0.2.0 backports_1.1.0 git2r_0.19.0
[9] magrittr_1.5 scales_0.4.1 evaluate_0.10.1 rlang_0.1.2
[13] stringi_1.1.5 lazyeval_0.2.0 labeling_0.3 tools_3.3.0
[17] stringr_1.2.0 munsell_0.4.3 yaml_2.1.14 colorspace_1.3-2
[21] htmltools_0.3.6 knitr_1.17 tibble_1.3.3 This R Markdown site was created with workflowr